Abstract
Cyclin-dependent kinase 5 (Cdk5) has been identified as a determinant of sensitivity to poly(ADP-ribose) polymerase (PARP) inhibitors. Here, the consequences of its depletion on cell survival, PARP activity, the recruitment of base excision repair (BER) proteins to DNA damage sites, and overall DNA single-strand break (SSB) repair were investigated using isogenic HeLa stably depleted (KD) and Control cell lines. Synthetic lethality achieved by disrupting PARP activity in Cdk5-deficient cells was confirmed, and the Cdk5KD cells were also found to be sensitive to the killing effects of ionizing radiation (IR) but not methyl methanesulfonate or neocarzinostatin. The recruitment profiles of GFP-PARP-1 and XRCC1-YFP to sites of micro-irradiated Cdk5KD cells were slower and reached lower maximum values, while the profile of GFP-PCNA recruitment was faster and attained higher maximum values compared to Control cells. Higher basal, IR, and hydrogen peroxide-induced polymer levels were observed in Cdk5KD compared to Control cells. Recruitment of GFP-PARP-1 in which serines 782, 785, and 786, potential Cdk5 phosphorylation targets, were mutated to alanines in micro-irradiated Control cells was also reduced. We hypothesize that Cdk5-dependent PARP-1 phosphorylation on one or more of these serines results in an attenuation of its ribosylating activity facilitating persistence at DNA damage sites. Despite these deficiencies, Cdk5KD cells are able to effectively repair SSBs probably via the long patch BER pathway, suggesting that the enhanced radiation sensitivity of Cdk5KD cells is due to a role of Cdk5 in other pathways or the altered polymer levels.
Similar content being viewed by others
Introduction
Poly(ADP-ribose) polymerase-1 (PARP-1) is a highly abundant early response sensor of DNA single-strand breaks (SSBs). It binds to SSBs and certain other forms of DNA strand breaks and catalyzes the transfer of poly(ADP-ribose) (PAR) chains from NAD+ onto itself and several other protein targets, generating a scaffold that is involved not only in DNA damage signaling and repair but also the modification of chromatin structure, transcription, RNA metabolism, cell death, and cell-cycle regulation (for review, see [1]). The absence or inhibition of PARP-1 has been shown in a variety of human cells and mouse models to result in a hypersensitivity to the cell-killing effects of DNA-damaging agents including ionizing radiation (IR) (see [2] and references therein). In addition, PARP inhibition in certain genetic backgrounds can result in hypersensitivity through a mechanism of synthetic lethality [3, 4]. Therefore, determining how PARP-1 is regulated is imperative for understanding not only its normal cellular functions but also its potential as a cancer therapeutic target.
Several studies in vitro and in vivo have suggested that PARP-1 activity itself may be regulated via phosphorylation at several sites and thus PARP-1 signaling in DNA repair pathways could also be modulated by phosphorylation. The kinases implicated include protein kinase C (PKC) α and β, extracellular signal-related kinases 1 and 2 (Erk1/2), c-Jun-N-terminal kinase 1 (JNK1), calmodulin kinase (CamK)-II, integrin-linked kinase (ILK), AMP-activated protein kinase (AMP-K), and cyclin-dependent kinase 5 (Cdk5) ([5] and references therein). A link between Cdk5 and PARP-1 was identified by Turner et al. [6] in a siRNA screen to identify kinases sensitizing cells to a PARP inhibitor. This serine/threonine kinase has distinct cellular roles as compared to other members of the large family of Cdks and is known to function in a neuronal cell context where it is essential for neuronal cell-cycle arrest and differentiation [7]. Turner et al. [6] showed that the Cdk5-silenced cells, in addition to an increased sensitivity to the cell-killing effects of PARP inhibitors, were also sensitive to the DNA-damaging agents camptothecin and cisplatin. Additionally, while Cdk5 silencing induced spontaneous formation of DNA double-strand breaks (DSBs) and markers of DSB repair, it was not required for early DSB signaling or DNA DSB repair. However, Cdk5 was found to be necessary for the activation of cell-cycle DNA-damage checkpoints and in particular the intra-S and G2/M cell-cycle checkpoints [6]. The mechanisms for these failed checkpoint activations are still not fully understood, but the background of greatly increased SSBs would be expected to lead to increased replication fork collapse and subsequent cell death.
In the present study, we have examined the impact of the stable depletion of Cdk5 on cell survival after exposure to the PARP inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) [8], methyl methanelsulfonate (MMS), neocarzinostatin (NCS), and IR. We have also investigated the impact of Cdk5 depletion on PARP activity, the recruitment of PARP-1, and other base excision repair (BER) proteins to sites of localized DNA damage and the overall capacity of Cdk5KD cells to repair IR-induced SSBs. The Cdk5-depleted cells displayed enhanced sensitivity to PARP inhibition confirming the results of Turner et al. [6], but of the panel of DNA-damaging agents tested, they only showed increased sensitivity to the cell-killing effects of IR compared to the response seen in the Control cells. These results suggest that there is an alteration in SSB processing in the Cdk5KD cells. Supporting this finding, we found that the persistence of GFP-tagged PARP-1 and YFP-tagged XRCC1 at sites of DNA damage was reduced in Cdk5KD cells and also that a PARP-1-GFP mutated at potential Cdk5 phosphorylation sites showed an altered DNA-damage recruitment profile in comparison to the Control cells. These results would suggest that Cdk5 modulates PARP-1’s activity and are supported by our finding that the Cdk5KD cells had higher basal and DNA damage-induced levels of polymer. Despite these differences in PARP-1 recruitment, the Cdk5KD cells were capable of religating all SSBs generated by IR, perhaps through a mechanism requiring PCNA, as the recruitment of GFP-tagged PCNA was found to be higher to localized damage sites in Cdk5KD cells compared to Control cells. These results suggest that the underlying molecular cause of the radiation sensitivity seen in the Cdk5KD cells is not the inability to repair either SSBs, nor DSBs generated directly, but may be due to the processing of IR-induced DNA damage in a replicating cell and the involvement of Cdk5 and/or PARP-1 in this process.
Materials and methods
Cell lines and gene silencing
shRNA sequences were designed with the DSIR program that also operates an exact similarity search algorithm for potential off-target detection [9]. Cloning in pEBVsiRNA vectors carrying a hygromycin B resistance cassette and establishment of stable knockdown and Control HeLa clones were carried out as previously described [10]. HeLa cells carrying the pBD650 plasmid that expressed an inefficient shRNA sequence were used as Controls [10]. The RNAi-targeted sequence for Cdk5 (NM_004935) was nucleotides 703–721 and was used to generate the Cdk5KD cells (clone 1499) used throughout this study, for PARP-1 (NM_001618) nucleotides 2,068–2,086 and for DNA-PKcs (NM_006904) nucleotides 5,980–5,998 [11]. Two additional Cdk5-depleted HeLa clones were generated where the RNAi-targeted sequences were nucleotides 455–473 generating clone 1,500, and nucleotides 41–59 generating clone 1,501. These two clones were used in the experiments presented in supplementary Fig. 4 only. All HeLa clones were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in the presence of 125 μg/ml hygromycin B under 5% CO2 in air. All tissue culture reagents were from Invitrogen.
Western blotting
Western blots of total cell extracts prepared using the M-PER reagent (Pierce, Perbio Science) containing a cocktail of protease and phosphatase inhibitors were carried out using precast NuPAGE® Novex 4–12% Bis–Tris gels (Invitrogen) as previously described [12]. The nitrocellulose membranes were blocked with either 5% bovine serum albumin or 5% nonfat milk (PARP-1 only) for 1 h at room temperature and then incubated with primary antibodies directed against PARP-1 (clone C2-10, BD Pharmingen, BD Biosciences) or Cdk5 (clone C-8, Santa Cruz) overnight at 4°C and with secondary antibodies coupled to horseradish peroxidase (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Membranes were probed with an enhanced chemiluminescence reagent and re-hybridized with α-tubulin antibody (clone DM1A, Sigma-Aldrich) for normalization.
Expression plasmids and transfection
pEGFP-NLS-PCNA, pEGFP-PARP-1, and XRCC1-pEYFP plasmids were kindly provided by Drs. P. Kannouche (FRE2939 CNRS, Institut Gustave-Roussy, Villejuif, France), V. Schreiber (UMR7175 CNRS, ESBS, Illkirch, France), K. W. Caldecott (Genome Damage and Stability Centre, University of Sussex, Brighton, UK), and P. Radicella (IRCM, CEA-DSV, Fontenay-aux-Roses, France), respectively. Plasmids were purified using a Macherey–Nagel NucleoBond® PC 500 EF kit. The three potential Cdk5 phosphorylation sites at serines 782, 785, and 786 (782-SDDSS-786) in PARP-1 were mutated in the pEGFP-PARP-1 plasmid to alanine residues (782-ADDAA-786) using the Stratagene QuikChange II XL site-directed mutagenesis kit PARP-1 S782-785-786A and the following primers:
-
PARP-1 S782-785-786A forward
-
5′-CTGCTCAGGGGAGGGGCTGATGATGCCGCCAAGGATCCCATCGATG-3′
-
PARP-1 S782-785-786A reverse
-
5′-CATCGATGGGATCCTTGGCGGCATCATCAGCCCCTCCCCTGAGCAG-3′.
The PCR conditions were: 95°C-2′; (95°C-1′, 55°C-1′, 65°C-10′) 20 cycles, 65°C-10′, and then a hold at 10°C until further processing, using a Veriti™ 96-well thermal cycler. The PARP-1 coding region of the mutated plasmid was resequenced before use in eight fragments (full details on request) using the BigDye® Terminator v1.1 cycle sequencing kit. Twenty-four hours before transfection, cells were plated at 50% confluence onto 25-mm round coverslips and transfected 48 h before the live-cell experiments using JET PEI (Polyplus transfection, Ozyme) according to the manufacturer’s recommendations.
Clonogenic cell survival assays
Control (1 × 103), PARP-1KD, DNA-PKcsKD, Cdk5KD (2 × 103) cells were plated in 25-cm2 flasks in triplicate, and allowed to adhere for 3–5 h at 37°C. For the inhibition of Cdk5 activity, cells were then grown for 24 h in the presence of 10 μM roscovitine (Sigma-Aldrich) in 1% DMSO, and for the inhibition of PARP activity cells for 24 h in the presence of 10 μM ABT 888 (Alexis Biochemicals) in 1% DMSO. For the assessment of cell survival after treatment with DNA damaging agents, cells were either treated with increasing concentrations of MMS (Sigma-Aldrich) for 1 h or NCS added directly to the medium. After all drug treatments, except NCS, which is rapidly degraded in tissue culture medium, cells were washed with Hank’s balanced salt solution and drug-free medium added. γ-Ray irradiation for the determination of clonogenic radiation survival was performed at room temperature in culture medium using an IBL-637 (137Cs) irradiator (CIS-Biointernational) at a dose rate of 0.70 Gy/min as previously described [12].
Colonies were allowed to grow for 10–15 days, before fixing with methanol, staining with Coomassie, and counting. Small colonies (<50 cells) were disregarded. The colony count relative to mock-irradiated cells (S) was adjusted for best fit to an exponential (Ln S = −α · D) (for the treatment of DNA-PKcsKD cells with NCS) or to the classical linear-quadratic equation (Ln S = −α · D−β · D 2) (all other treatments) where D is the treatment dose and α and β adjustable parameters characterizing the response. Calculations were made through non-linear least-squares regression taking all data points into account, using Kaleidagraph software (Synergy Software).
Live-cell videomicroscopy
Live-cell videomicroscopy was carried out as previously described [12]. DNA photodamage was generated using a 405-nm laser line (diode) set to maximum output for 500 ms and focused onto a single spot of constant size (176 nm) within the nucleus to generate a point of photodamage with a constant amount of energy. Recruitment of the protein of interest was then monitored by fluorescence using the same setting as for the pre-damage sequence. Nomarski contrast images were taken in parallel. Images were captured at intervals of 2–5 s.
Photodamage kinetics were analyzed under the ImageJ software (Rasband, W.S., ImageJ, U.S. National Institutes of Health, http://imagej.nih.gov/ij/, 1997–2011) using an in-house developed macro. Data extraction is a semi-automated process where the user is first invited to draw two regions of interest (ROI): a nuclear region and the site of photodamage. The background ROI is automatically set to the inverse of the nuclear ROI. For each time point, the integrated intensity is extracted from the three ROIs. Intensity from the photodamage site is corrected for both the observational photobleaching and background, and normalized so that its intensity at start-up equals 1: corrected intensity = ((focus intensity (t)−background intensity(t) × focus area/background area) × nuclear intensity(t))(focus intensity(t 0) × nuclear intensity(t 0). The timepoint of induction of the photodamage is extracted from the normalized curve by determining when the maximum change in intensity is achieved between two successive images. The normalized time course from the damage on was fitted to a mono-exponential recovery of the form y = a (1−exp(−b × t) + c, which allows the estimation of the half-time maximum recruitment (t 1/2 = −ln((a + c)/2 × a)/b) as well as the expected maximum recruitment (max = a + c).
Detection of PAR formation by immunofluorescence
For the detection of polymer formation after exposure to IR, cells were plated onto glass coverslips in six-well plates and 48 h later were exposed to 8 Gy delivered over 3 min. Three min after the end of the irradiation, cells were fixed (4% formaldehyde in PBS for 25 min). Cells were then washed three times with PBS containing 0.1% Triton X-100, rinsed with PBS alone, and re-washed once with PBSMT (PBS with 2% milk, 0.5% Triton X-100). After blocking (30 min, room temperature in PBSMT) cells were incubated with a primary antibody against poly(ADP-ribose) polymers (Trevigen, clone 10HA, 1/400 dilution in PBSMT) for 1 h at 37°C. For the detection of PAR formation after exposure to H202, the growth medium of the plated cells was replaced by PBS alone or by PBS containing 1 mM H202 (Sigma-Aldrich). After a 10-min treatment and a wash with PBS, cells were fixed (2% paraformaldehyde, 0.1% Triton X-100 in PBS for 20 min). Cells were then washed three times with PBS containing 0.1% Triton X-100 and once with TBSAT (TBS with 1% BSA, 0.5% Triton X-100) and then incubated with the same primary antibody as above. Following washing, the slides were incubated with AlexaFluor 488 goat anti-rabbit IgG (Molecular Probes, 1/500 dilution for 1 h at RT). After washing, coverslips were mounted with Vectashield containing DAPI 1.5 μg/ml (Vector Laboratories). Images of fields were acquired on a Leica confocal SP5 system, with a 63× objective (line average of four). At least 200 cells were analyzed per experimental condition. The global fluorescence intensity by nucleus was quantified in arbitrary units (AU) using a macro with the Image J software.
Measurements of NAD(P)H levels
Changes in the intra-cellular levels of NAD(P)H following H2O2 treatment were monitored as previously described using the Cell Counting Kit-8 (CCK-8) solution (Dojindo Laboratories) [13]. Briefly, 1 × 104 cells were plated in a 96-well plate in 100 μl of growth medium. The next day, the cells were treated with a range of H2O2 doses, in the presence of 10 μl of CCK-8 solution. The absorbance at 450 nm was measured 4 h after the start of treatment and the changes in NAD(P)H levels were calculated as a percentage of the absorbance in the absence of H2O2 treatment. Measurements were done in quadruplate.
Measurements of PARG activity
PARG activity was measured in cell extracts using a commercially available kit (HT Chemiluminescent PARG Assay Kit, Trevigen) according to the manufacturers’ instructions. Briefly, the relative PARG activity in cell extracts was measured by quantification of the loss of biotinylated PAR from histones attached to strip wells in a 96-well format.
Analysis of SSB repair by alkaline filter elution
Radiation-induced SSBs were measured by alkaline filter elution over polycarbonate filters as previously described [12] based on the technique of Filipski and Kohn [14], which allows the titration of direct SSBs, alkali-labile and abasic sites altogether. In brief, cells were grown in the presence of [2-14C] thymidine (0.05 μCi/ml) (Perkin-Elmer) for 36 h. The medium was then replaced and the cells were returned to the incubator for at least 2 h before exposure to 5 Gy gamma rays at a dose rate of 2.64 Gy/min, at room temperature. Cells were returned to the incubator for fixed times to allow strand break repair to occur. For the determination of the initial yield of SSBs, irradiation was performed on ice to prevent SSB rejoining. At the end of the incubation period, the flasks were rapidly rinsed once with ice-cold PBS-10 mM EDTA, and maintained on ice for not more than 5 min before harvesting with the aid of a cell scraper. An amount of 8 × 105 cells were deposited onto polycarbonate filters, then lysed, and the DNA fragments were eluted and counted. Each measurement was performed in duplicate. The results shown in Fig. 5 were fitted to the exponential equation,
where R, R 0, R ∞ are the percentages of DNA retained on filters at time t, at time zero and at completion of the reaction, respectively.
Statistics
A non-parametric Mann–Whitney U test was performed using StatEL (AD Science, Paris, France) on paired data sets to assess differences between survival curves (Fig. 1). All data presented in the other figures was analyzed with Prism GraphPad 5.0b software (GraphPad Software Inc.) using unpaired t tests with p being the calculated error risk.
Clonogenic cell survival of Cdk5-depleted HeLa cells. a Western-blot analysis confirming stable depletion of Cdk5 or PARP-1 expression in Cdk5 KD and PARP-1KD cells, respectively. b Clonogenic cell survival of Control or Cdk5KD cells after exposure to DMSO or the PARP-1 inhibitor ABT888 (10 μM, 24 h). c Clonogenic cell survival of Control or PARP-1KD cells exposed to DMSO or the Cdk5 inhibitor roscovitine (10 μM, 24 h). Data represents mean ± SD, for at least two independent experiments in triplicate for all conditions **p < 0.004
Results
The loss of Cdk5 expression results in increased sensitivity to PARP inhibition and the killing effects of IR
In order to investigate whether Cdk5 depletion has a direct impact on PARP-1 activity and sensitivity to a panel of DNA-damaging agents, we established a stably depleted Cdk5 HeLa cell line (Cdk5KD) using an RNAi system that targeted nucleotides 703–721. These Cdk5KD cells (clone 1,499) were cultivated for more than 80 days in culture with levels of the targeted proteins remaining below 30%, as assessed by the quantification of Western blots, of that seen in Control cells (Fig. 1a). A similar low level of Cdk5 protein expression was seen in two additional Cdk5-depleted clones, 1,500 and 1,501, in which nucleotides 455–473 and 41–59 were targeted, respectively (supplementary Fig. 4f).
The depletion of Cdk5 resulted in a significant increase in the sensitivity to cell killing following treatment with the PARP inhibitor ABT888 for 24 h compared to the response seen in Control cells under the same experimental conditions (p < 0.004) (Fig. 1b). In parallel experiments, comparing the survival of Control and HeLa cells depleted for PARP-1 cells after treatment with 10 μM of roscovitine for 24 h, an inhibitor of Cdk activity, a significantly lower survival was seen in the PARP-1KD cells (p < 0.002) (Fig. 1c). These results confirm those of Turner et al. [6] that synthetic lethality occurs when PARP and Cdk5 activity are both disrupted in stably depleted non-neuronal cell lines and lends support to a model that the DNA damage response is modified when both activities are compromised.
In order to further characterize the Cdk5KD cells, we compared the killing effects of a panel of three DNA-damaging agents, MMS, NCS, and IR, in these cells to that seen in unsynchronized cell populations of Control cells or cells invalidated for PARP-1 or DNA-PKcs. The different combinations were chosen to generate a spectrum of DNA lesions in backgrounds where different repair pathways are compromised. The base damage formed after exposure to MMS will either be eliminated from the DNA by spontaneous depurination or will be actively repaired through the action of glycosylases such as 3-methyladenine glycosylase and then through the later stages of the BER pathway [15]. NCS is a complex consisting of a dodecadiyne antibiotic (NCSChrom) reversibly bound to a carrier protein. NCSChrom cleaves DNA through a suicide reaction, leaving no residual active drug after a few minutes of incubation. The major DNA lesions induced by NCSChrom in DNA result from radical attack [16] and consist of a blunt end break with a thymidine-5′-aldehyde residue on one strand, with an atypical abasic site at two-nucleotide interval on the complementary strand. This NCS-induced damage is rapidly converted into DSBs in living cells and thus NCS is considered to be a DSB-inducing agent. Finally, IR generates a variety of DNA base adducts, SSBs, and DSBs, as well as complex DNA lesions.
Cdk5KD and PARP-1KD cells were found to be approximately 1.6 times more sensitive to the killing effects of IR than Control cells, based on the surviving fraction at 2 Gy (SF2) (Fig. 2a). No increased sensitivity was seen in either the Cdk5KD cells or the PARP-1KD cells to either MMS or NCS. However, the DNA-PKcsKD cells were extremely sensitive to NCS (Fig. 2b, c). These similarities in cell survival between the Cdk5KD and the Control cells after treatment with MMS or NCS would suggest that the methylated DNA adducts and DSBs generated directly in the DNA after such treatments can be repaired using mechanisms that do not require Cdk5 but would suggest that Cdk5 is involved in the repair of SSBs, a process which involves PARP-1’s catalytic activity.
Clonogenic cell survival to IR, NCS, and MMS of Cdk5KD cells. a IR survival in Control, PARP-1, and Cdk5-depleted cells. α = 0.363 ± 0.019 Gy−1, β = 0.025 ± 0.007 Gy−2 for Control cells; α = 0.446 ± 0.042 Gy−1, β = 0.105 ± 0.025 Gy−2 for PARP-1KD cells; α = 0.519 ± 0.019 Gy−1, β = 0.056 ± 0.011 Gy−2 for Cdk5KD cells. b Survival to NCS, and c MMS. Data represents mean ± SD of 2–4 independent experiments with each dose in triplicate
Cdk5 depletion modulates PARP-1 recruitment to sites of localized DNA damage and its enzymatic activity
In order to examine whether the depletion of Cdk5 had an impact on the recruitment of PARP-1, and its partner XRCC1, we made use of the technique of laser micro-irradiation using a 405-nm laser to generate localized DNA damage in the nucleus of the cells transiently expressing GFP or YFP tagged PARP-1 or XRCC1, respectively. Confocal microscopy was used to follow and quantify the recruitment and persistence of these proteins based on the increase in fluorescence at the site of DNA damage relative to the background fluorescence of the nucleus, reported as the relative spot intensity.
The time-course of the recruitment of GFP-PARP-1 to damage sites seen in the Control cells was extremely rapid, as previously reported [12, 17–20]. However, in the Cdk5KD cells whilst GFP-PARP-1 was detected at damage sites immediately after their formation (Fig. 3a, first and second rows), the quantification of the relative spot intensity with time showed a statistically reduced level (p < 10−3) of GFP-PARP-1 at the damage sites in the Cdk5KD cells compared to Control cells from 50 s post-irradiation and which persisted up to 300 s post-irradiation (Fig. 3b). A statistically different (p < 10−3) profile of GFP-PARP-1 recruitment was also seen in the Cdk5 depleted clones 1,500 and 1,501 compared to the Control cells (supplementary Fig. 4a and b). As the three serine residues at amino acids 782, 785 and 786 in PARP-1 have been previously reported as in vitro target sites for phosphorylation by Cdk5 [5], we generated a mutant GFP-tagged PARP-1 plasmid in which all three were mutated to alanines (PARP-1 MUT-GFP). Under the same experimental conditions, we found that this mutated form of PARP-1 was also recruited to sites of DNA damage in Control cells immediately following micro-irradiation (0 min), however, its accumulation and persistence was less than that of the control GFP-PARP-1 and showed no statistical difference to the recruitment profile of GFP-PARP-1 in the Cdk5KD cells. (Fig. 3a rows 2 and 3 and 3b). These results would suggest that the phosphorylation of PARP-1 via Cdk5's kinase activity is necessary for its persistence at damage sites.
Recruitment of SSBR proteins in Control and Cdk5KD cells to laser damage sites. a Representative images of the recruitment of fluorescent-tagged PARP-1, a serine-mutated PARP-1 (PARP-1 MUT), or XRCC1 to DNA damage sites in Control or Cdk5KD cells. Scale bar: 5 μm. b Quantitative analysis of the relative spot intensity with time of GFP-PARP-1 and PARP-1 MUT-GFP recruitment in Control cells and GFP-PARP-1 in Cdk5KD cells and of c XRCC1-YFP recruitment in Control and Cdk5KD cells with time. Data represents mean relative spot intensity ± SEM, n = 7–12 individual cells from three independent experiments for GFP-PARP-1 and XRCC1-YFP experiments and two independent experiments for PARP-1 MUT-GFP experiments
It has been well-documented that XRCC1 is recruited to local sites of DNA damage in a PAR and PARP-1-dependent fashion [12, 19, 21, 22]. Therefore, in order to verify the consequences of the reduced persistence of PARP-1 at sites of micro-irradiation-induced DNA damage sites in Cdk5KD cells, we next investigated the recruitment of XRCC1-YFP under the same experimental conditions (Fig. 3a). In agreement with other published studies using a similar experimental set-up, a rapid recruitment of XRCC1 was seen in the Control cells [12, 19, 21–23]. However, while we found that the relative intensity of XRCC1-YFP immediately after micro-irradiation (0 min) was not affected by the loss of Cdk5 expression (Fig. 3a rows 4, 5 and 3c), its accumulation was statistically lower (p < 10−4) in Cdk5KD cells at time points from 100 s after damage formation as compared to Control cells (Fig. 3c). This result was confirmed in the two additional Cdk5-depleted clones 1,500 and 1,501 (supplementary Fig. 4c and d). Supporting these findings, we observed a similar decreased level and slower accumulation of fluorescence from the YFP-tagged XRCC1 in Control cells exposed to the Cdk5 inhibitor roscovitine (supplementary Fig. 1).
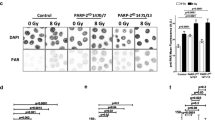
The persistence of PARP-1 at sites of SSBs is regulated by the PAR chains formed during the automodification of the PARP-1 protein itself. These PAR chains impart a negative charge on PARP-1 and result in its eventual dissociation from the DNA as a consequence of charge repulsion between the negative charge of the DNA and the PAR chains [1, 22]. In order to investigate whether the reason for the reduced accumulation of PARP-1 observed in the Cdk5KD cells could be related to differences in the levels of PAR formed in the two cell types, we measured the basal levels and the levels formed in response to DNA damage induced by IR or H202 using immunofluorescence making use of a specific anti-PAR antibody.
A statistically higher basal level of PAR was detected in the Cdk5KD cells and also in the clones 1,500 and 1,501 compared to the Control cells (Fig. 4b, d and supplementary Fig. 4e). After exposure to IR, the PAR levels were increased in both cell types with significantly higher absolute levels being found in the Cdk5KD compared to the Control cells (p < 10−4, n = 300–500 cells) (Fig. 4a, b). A similar response was seen after exposure to 1 mM H2O2 (p < 10−5, n = 400–800 cells) (Fig. 4c, d). Confirmatory data obtained from clones 1,500 and 1,501 showing significantly higher absolute PAR levels compared to Control cells after treatment with IR is shown in supplementary Fig. 4e (p < 10−5, n = 200–300 cells).
Quantification of poly(ADP-ribose) (PAR) formation. Representative images of PAR immunofluorescence in Control cells or Cdk5KD cells before and after exposure to 8 Gy of ionizing radiation (a, zooming set to 4) or after treatment with 1 mM H2O2 (c, zooming set to 3). Scale bar: 10 μM. b, d Quantification of the PAR staining. Data represents the mean ± SD *** (p < 10−4) based on the fluorescence intensity in over 300 cells/treatment group in 2–4 independent experiments
These observations of higher levels of PAR formation were validated by measuring the changes in NAD(P)H levels using a colorimetric assay [13]. However, due to the technical limitations associated with the assay, this could only be done after the H2O2 treatment, where the exposure could be carried out over a longer period of time. While the PARP-1KD cells showed a negligible decrease in NAD(P)H levels under these experimental conditions, the Cdk5KD cells showed a significantly greater consumption of NAD(P)H compared to the Control cells (p < 10−4), indicative of a higher PARP activity after DNA damage induced by H2O2 (supplementary Fig. 2).
A possible explanation for the finding of higher polymer formation in Cdk5KD cells is that the activity of poly(ADP-ribosyl) glycohydrolase (PARG), responsible for removing the PAR polymers from PARP-1 [24], is reduced in the absence of Cdk5. Post-translational modifications of PARG have been reported at a number of positions, several of which are located in the N-terminal putative regulatory region raising the possibility that as for PARP-1, phosphorylation events may modulate PARG activity [5, 25, 26]. This N-terminal region contains a sequence around Ser 137 that shows a high degree of homology with the consensus sequence for Cdk5 phosphorylation (KS/TPXK) [27]. However, no differences were seen between the Cdk5KD and Control cells in either their PARG mRNA or protein levels (data not shown), nor in either the basal PARG activity or after exposure of the cells to IR (supplementary Fig. 3). Based on these results and the recruitment data, we hypothesize that the phosphorylation of the PARP-1 protein by Cdk5 on one or more of the serines 782, 785, and 786 results in an attenuation of its ribosylating activity facilitating its persistence at the sites of DNA damage.
Cells lacking Cdk5 are able to effectively repair DNA single-strand breaks (SSB)
In order to investigate whether the impaired recruitment of PARP-1 and XRCC1 in the Cdk5KD cells had an impact on SSB rejoining, the kinetics was compared in unsynchronized Control and Cdk5KD cells after exposure to gamma irradiation (5 Gy) using alkaline elution. Immediately following irradiation, as little as 5% of the total [2-14C] thymidine-labeled DNA from lysed Control and Cdk5KD cells was retained on the filters and in a time-dependent manner over 1 h, the SSB repair proceeds to completion with nearly 100% of the DNA retained on the filter, indicating complete SSB rejoining, by this time point in both cell lines (Fig. 5).
Alkaline elution analysis of the kinetics of SSB rejoining in Control and Cdk5KD cells. The curves were drawn to an exponential equation (see “Materials and methods”) for best fit to experimental data α = 0.109 ± 0.050 min−1 (t 1/2 = 6.36 min), R ∞ = 95.35% (Control); α = 0.087 ± 0.050 min−1 (t 1/2 = 8.01 min), R ∞ = 96.89% (Cdk5KD). Data represents mean and SD for two independent experiments. Where error bars are missing, they were smaller than the size of the symbols
DNA SSB repair can also proceed through the alternative long patch (LP) BER sub-pathway involving the binding of PCNA to sites of DNA damage. In order to investigate whether this mechanism could contribute to the repair of SSBs in the Cdk5KD cells, GFP-tagged PCNA was transiently transfected into these and Control cells, and its recruitment to DNA-damage sites produced by laser micro-irradiation were assessed as previously described [12, 18, 20, 21]. In the Control cells, the fluorescence intensity of PCNA at the irradiated site was significantly lower than for XRCC1, which is in good agreement with earlier studies [12, 18, 23], and increased slowly over the observation period. However, in the Cdk5KD cells, the amount and rate of PCNA-GFP accumulation was significantly higher (Fig. 6).
Analysis of GFP-tagged PCNA recruitment to laser damage sites in Control or Cdk5KD cells. a Representative images of Control and Cdk5KD cells expressing PCNA-GFP before irradiation (Pre-IR), immediately following irradiation (0 min) and after 5 min. Scale bar (lower right) is 5 μm. b Quantification of spot diameter of PCNA-GFP at sites of micro-irradiation in Control and Cdk5KD at 5 min. Data represents mean ± SEM, n = 9–10 individual cells from two independent experiments, *p < 0.05. c To quantify the kinetics of recruitment of these proteins to DNA damage sites, images were collected over a time course from 0 to 250 s every 5 s. Data represents the mean ± SEM, n = 9–10 individual cells from one representative experiment
These results suggest that the SSBs generated might also be repaired via a PCNA-dependent pathway compensating for the deficiencies in the recruitment of the PARP-1 and XRCC1 in the Cdk5KD cells. Thus, although the loss of Cdk5 does not prevent cells from resolving SSBs, it does have an impact on PARP-1 activity and radiation sensitivity.
Discussion
While deficiencies in DNA repair processes are associated with cancer susceptibility and tumor resistance to different DNA-damaging cancer therapies, the inhibition of DNA repair by small-molecule inhibitors can potentially be exploited to sensitize tumor cells in certain genetic backgrounds or when used in combination with chemo and radiotherapy. PARP-1 is one such target and based on the compelling evidence that defects in DSB repair in breast cancer, particularly in BRCA1 and BRCA2 backgrounds, sensitize cells to PARP inhibitors, clinical trials are now currently being conducted with PARP inhibitors in various cancer types (see [28] for recent review). Since these landmark studies, there has been a focus on identifying additional indirect therapeutic targets that modulate PARP activity and could also increase the efficacy of combination cancer therapies. Cdk5 was identified as such a candidate given that its silencing confers significant sensitization to PARP inhibitors [6] and its potential role in DNA repair mechanisms [29, 30]. Here, we confirm that cells with depleted Cdk5 expression are highly sensitive to PARP inhibition and present data that in three independent clones in which different regions of the Cdk5 mRNA were targeted, the subsequent depletion of Cdk5 protein levels alters PARP-1’s recruitment and persistence at sites of DNA damage and PAR formation is increased after exposure to DNA-damaging agents. These results taken together with the finding that the recruitment profile of GFP-tagged PARP-1 in which the serines 782, 785, and 786, reported to be Cdk5 phosphorylation targets in vitro [5], are replaced with alanine residues to the sites of DNA damage in Control cells is the same as that of GFP-tagged PARP-1 in the Cdk5KD cells, would suggest that Cdk5 might modify the PARP-1 protein. The altered recruitment could formally be a consequence of a conformational change brought about by the substitution of the serines at these positions by alanines. However, we do not believe this to be likely based on an analysis of PARP-1 structures available in public databases that show that these residues are located within a flexible and highly accessible external loop of the PARP-1 protein (Saettel and Pennaneach, unpublished data). Based on all our experimental data and this consideration, it would appear that the post-translational modification of PARP-1 by Cdk5 is necessary for its maximal recruitment to DNA-damage sites and thus for normal PARP-1 function in SSBR and as a consequence, also the recruitment and persistence of XRCC1 at damage sites.
The regulation of PARP-1 activity by phosphorylation has been described in vitro and in vivo. For instance, Aoufouchi and Shall [31] demonstrated that during the maturation of Xenopus laevis oocytes to unfertilized eggs, PARP-1 is phosphorylated, which switches on its enzymatic activity, while its phosphorylation by protein kinase C in vitro was found to inhibit PARP activity and its binding capacity [32, 33]. Additionally, it has been found in vitro that the dephosphorylation of recombinant human PARP-1 with calf–intestinal alkaline phosphatase reduces its enzymatic activity by about 75% [34]. The post-translational modification of PARP-1 is clearly a complex process impacting on its activity. Several potential phosphorylation sites within the PARP-1 protein have been identified using bioinformatic approaches and mass spectrometry analysis that implicate multiple kinases, some capable of phosphorylating several residues within the protein. Phosphorylation of some of these sites has been confirmed in vitro using kinases such as PKCβ, ERK1/2, JNK1, CamK-II, and CKII, in addition to Cdk5 [5, 33–36]. The impact of phosphorylation at specific sites on PARP-1 function has not, however, been extensively studied. Gagne et al. [5] analyzed the recruitment kinetics of GFP-tagged phospho-mutants within the first N-terminal zinc finger domain of PARP-1 that is required for DNA binding. They found that a glutamate substituted Ser 41, which mimics a permanently phosphorylated protein, impaired both the recruitment and the subsequent persistence of PARP-1 at laser micro-irradiation-induced damage sites while similar substitutions to mimic phosphorylated residues at Ser 27 and 32 had little effect. This result is in contrast to our findings where the absence of Cdk5 or mutating the serines 782, 785, and 786 within PARP-1 that have been shown to be phosphorylated in vitro by Cdk5 [5] are associated with increased intra-cellular polymer levels and lower recruitment of PARP-1 to DNA-damage sites. As discussed above, this altered PARP-1 profile under these experimental conditions could be explained by either an increased catalytic activity of PARP-1 in the absence of Cdk5, which would result in an excessive automodification of PARP-1, itself a major target of the polymers formed, or thus its dissociation from the DNA through charge repulsion or alternatively a decreased PARG activity. Our experimental results support the first option, as no differences in PARG activity were found between Cdk5KD and Control cells.
One phenotype of the Cdk5KD cells that remains to be fully explained is their sensitivity to the killing effects of IR. Whether this observation is just a consequence of the altered PARP-1 activity, which would result in a perturbation of polyADP ribosylation in the Cdk5KD cells, or is due to the involvement of Cdk5 in other DNA repair and/or other pathways such as cell cycle control, through post-translational modifications or transcriptional control, needs to be established. The role of Cdk5 in cell-cycle control has been extensively studied in the context of neuronal cells where Cdk5 is associated with a fatal re-entry of post-mitotic cells into the cell cycle [7, 37–39], and as discussed above, Turner et al. [6] showed a defect in intra-S and G2/M checkpoints following treatment with 10 Gy of IR in proliferating Cdk5-deficient cells. In addition, Cdk5 has been shown to phosphorylate the STAT3 protein following DNA damage induced by the inhibition of topoisomerase I [40, 41] and other reports have shown that Cdk5 can directly phosphorylate key players in BER and DSB repair, such as Ape1 or ATM and alter their activity [29, 30]. Based on our data, it is tempting to speculate that the increased radiation sensitivity is not due to the lack of repair of SSBs per se, as under the experimental conditions used, a complete religation of the SSBs generated after exposure to IR is seen in these cells with a kinetic profile that resembles that seen in Control cells. This may involve a PCNA-dependent sub-pathway compensating for the altered recruitment of PARP-1 and XRCC1 in the Cdk5KD cells. However, there is no experimental evidence that would suggest that a switch between SP and LP BER sub-pathways is related to increased radiation sensitivity. The observations that no differences were observed in the cell survival between the Cdk5KD and Control cells after treatment with NCS, an agent that directly produces DNA DSBs, would suggest that the detection and repair of DSBs is not associated either with the increased radiation sensitivity but would support the hypothesis that the handling of SSBs, perhaps at the replication fork, and the activation of the necessary cell-cycle checkpoints, underlies this radiation sensitivity. It is interesting to note that Turner et al. [6] reported that their Cdk5-depleted cells were sensitive to the cell-killing effects of camptothecin, which blocks topoisomerase 1 and whose toxicity is linked to the conversion of SSBs to DSBs when the replication fork collides with complexes formed by DNA and camptothecin [42]. An alternative possibility could be a more indirect effect of the Cdk5 depletion and one linked to the modification of PARP activity in the Cdk5-depleted cells. In addition to its role in SSB repair, in response to DNA damage, PARP-1 poly(ADP-ribosyl)ates the histones situated around the site of the damage, facilitating the relaxation of the chromatin (for a review see [43]). The higher levels of PAR seen in the Cdk5-depleted clones both before and after exposure to IR, and also H202, could cause excessive or prolonged chromatin relaxation providing an additional, potentially additive mechanism of radiation-induced cell death in these cells.
In conclusion, several studies have now demonstrated a role for Cdk5 in cell metabolism in non-neuronal cell types and more specifically that Cdk5 can directly modify proteins integral in the DNA damage/DNA repair response pathways. A depletion in the level of Cdk5 results in an increased level of PAR polymer formation and an altered profile of PARP-1 recruitment at damaged sites but not an overall deficit in SSB repair. Turner et al. [6] reported a genomic loss of CDK5 in 5.5% of breast cancers and taken together with our results suggest that CDK5 gene might represent a potential biomarker for a specific sub-population of breast cancer patients that would benefit from combined PARP-1 inhibitor and radiation treatment. Additionally, modification of the Cdk5 activity in tumor cells could be a novel therapeutic target in conjunction with this combination treatment.
References
Schreiber V, Dantzer F, Ame JC, de Murcia G (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7(7):517–528
Noel G, Godon C, Fernet M, Giocanti N, Megnin-Chanet F, Favaudon V (2006) Radiosensitization by the poly(ADP-ribose) polymerase inhibitor 4-amino-1, 8-naphthalimide is specific of the S phase of the cell cycle and involves arrest of DNA synthesis. Mol Cancer Ther 5(3):564–574
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434(7035):913–917
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434(7035):917–921
Gagne JP, Moreel X, Gagne P, Labelle Y, Droit A, Chevalier-Pare M, Bourassa S, McDonald D, Hendzel MJ, Prigent C, Poirier GG (2009) Proteomic investigation of phosphorylation sites in poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase. J Proteome Res 8(2):1014–1029
Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, Rayter S, Tutt AN, Ashworth A (2008) A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J 27(9):1368–1377
Cicero S, Herrup K (2005) Cyclin-dependent kinase 5 is essential for neuronal cell cycle arrest and differentiation. J Neurosci 25(42):9658–9668
Penning TD, Zhu GD, Gandhi VB, Gong J, Liu X, Shi Y, Klinghofer V, Johnson EF, Donawho CK, Frost DJ, Bontcheva-Diaz V, Bouska JJ, Osterling DJ, Olson AM, Marsh KC, Luo Y, Giranda VL (2009) Discovery of the poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem 52(2):514–523
Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y (2006) An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics 7:520
Biard DS, Despras E, Sarasin A, Angulo JF (2005) Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: application to NER gene silencing. Mol Cancer Res 3(9):519–529
Despras E, Pfeiffer P, Salles B, Calsou P, Kuhfittig-Kulle S, Angulo JF, Biard DS (2007) Long-term XPC silencing reduces DNA double-strand break repair. Cancer Res 67(6):2526–2534
Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V (2008) PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res 36(13):4454–4464
Nakamura J, Asakura S, Hester SD, de Murcia G, Caldecott KW, Swenberg JA (2003) Quantitation of intracellular NAD(P)H can monitor an imbalance of DNA single strand break repair in base excision repair deficient cells in real time. Nucleic Acids Res 31(17):e104
Filipski J, Kohn KW (1982) Ellipticine-induced protein-associated DNA breaks in isolated L1210 nuclei. Biochim Biophys Acta 698(3):280–286
Beranek DT, Weis CC, Swenson DH (1980) A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis 1(7):595–606
Dedon PC, Jiang ZW, Goldberg IH (1992) Neocarzinostatin-mediated DNA damage in a model AGT. ACT site: mechanistic studies of thiol-sensitive partitioning of C4′ DNA damage products. Biochemistry 31(7):1917–1927
Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG (2008) Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36(22):6959–6976
Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A (2004) In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci USA 101(38):13738–13743
Mortusewicz O, Ame JC, Schreiber V, Leonhardt H (2007) Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res 35(22):7665–7675
Mortusewicz O, Fouquerel E, Ame JC, Leonhardt H, Schreiber V (2011) PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res [Epub ahead of print]
Okano S, Lan L, Caldecott KW, Mori T, Yasui A (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol 23(11):3974–3981
Quenet D, El Ramy R, Schreiber V, Dantzer F (2009) The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol 41(1):60–65
Mortusewicz O, Leonhardt H (2007) XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol Biol 8:81
Ame JC, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, Schreiber V (2009) Radiation-induced mitotic catastrophe in PARG-deficient cells. J Cell Sci 122(12):1990–2002
Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA 101(33):12130–12135
Villen J, Beausoleil SA, Gerber SA, Gygi SP (2007) Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA 104(5):1488–1493
Lalioti V, Pulido D, Sandoval IV (2010) Cdk5, the multifunctional surveyor. Cell Cycle 9(2):284–311
Annunziata CM, O’Shaughnessy J (2010) Poly(ADP-ribose) polymerase as a novel therapeutic target in cancer. Clin Cancer Res 16(18):4517–4526
Huang E, Qu DAB, Zhang Y, Venderova K, Haque ME, Rousseaux MWC, Slack RS, Woulfe JM, Park DS (2010) The role of Cdk5-mediated apurinic/apyrimidinic endonuclease 1 phosphorylation in neuronal death. Nat Cell Biol 12(6):563–571
Tian B, Yang Q, Mao Z (2009) Phosphorylation of ATM by Cdk5 mediates DNA damage signalling and regulates neuronal death. Nat Cell Biol 11(2):211–218
Aoufouchi S, Shall S (1997) Regulation by phosphorylation of Xenopus laevis poly(ADP-ribose) polymerase enzyme activity during oocyte maturation. Biochem J 325(Pt 2):543–551
Bauer PI, Farkas G, Buday L, Mikala G, Meszaros G, Kun E, Farago A (1992) Inhibition of DNA binding by the phosphorylation of poly ADP-ribose polymerase protein catalysed by protein kinase C. Biochem Biophys Res Commun 187(2):730–736
Tanaka Y, Koide SS, Yoshihara K, Kamiya T (1987) Poly(ADP-ribose) synthetase is phosphorylated by protein kinase C in vitro. Biochem Biophys Res Commun 148(2):709–717
Kauppinen TM, Chin WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA (2006) Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci USA 103(18):7136–7141
Ullrich O, Diestel A, Eyupoglu IY, Nitsch R (2001) Regulation of microglial expression of integrins by poly(ADP-ribose) polymerase-1. Nat Cell Biol 3(12):1035–1042
Walker JW, Jijon HB, Madsen KL (2006) AMP-activated protein kinase is a positive regulator of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun 342(1):336–341
Yang Y, Geldmacher DS, Herrup K (2001) DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci 21(8):2661–2668
Yang Y, Mufson EJ, Herrup K (2003) Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer’s disease. J Neurosci 23(7):2557–2563
Zhang J, Cicero SA, Wang L, Romito-Digiacomo RR, Yang Y, Herrup K (2008) Nuclear localization of Cdk5 is a key determinant in the postmitotic state of neurons. Proc Natl Acad Sci USA 105(25):8772–8777
Fu AK, Fu WY, Ng AK, Chien WW, Ng YP, Wang JH, Ip NY (2004) Cyclin-dependent kinase 5 phosphorylates signal transducer and activator of transcription 3 and regulates its transcriptional activity. Proc Natl Acad Sci USA 101(17):6728–6733
Mullaart E, Boerrigter ME, Ravid R, Swaab DF, Vijg J (1990) Increased levels of DNA breaks in cerebral cortex of Alzheimer’s disease patients. Neurobiol Aging 11(3):169–173
Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, Kohn KW (2003) Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res 532(1–2):173–203
Rouleau M, Aubin RA, Poirier GG (2004) Poly(ADP-ribosyl)ated chromatin domains: access granted. J Cell Sci 117(Pt 6):815–825
Acknowledgments
We wish to thank Camille Godon, Celia Rosilio, and Yun-Han Huang who participated in early stages of this project, Amélie Croset who shared PAR protocols with us, and Nicolas Saettel for helpful discussions on PARP structures. The financial support of Electricité de France, INSERM, and Institut Curie are gratefully acknowledged. Celeste Bolin and Tomasz Zaremba were supported by Institute Curie’s International Postdoctoral fellow program and Tomasz Zaremba was also supported by a fellowship from the Fondation Pierre-Gilles de Gennes.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors Celeste Bolin and Mohammed-Tayyib Boudra contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2011_811_MOESM1_ESM.tif
Supplemental Fig. 1 XRCC1-YFP recruitment at sites of micro-irradiation in Control cells. a Representative images of the recruitment of XRCC1-YFP in Control cells treated with or without 10 μM of the Cdk5 inhibitor roscovitine for 24 h prior to micro-irradiation. b Quantification of the recruitment of XRCC1-YFP over a time course from 0 to 250 s. Scale bar (lower right) is 5 μm. Data represents mean and SEM, n = 3–10 individual cells from two independent experiments. (TIFF 1113 kb)
18_2011_811_MOESM2_ESM.tif
Supplemental Fig. 2 NAD(P)H levels in Control and Cdk5KD cells after treatment with a range of H2O2 concentrations, expressed as average percentages of non-treated levels. Error bars represent the SD of three independent experiments, ***p < 10−4. (TIFF 329 kb)
18_2011_811_MOESM3_ESM.tif
Supplemental Fig. 3 Effects of Cdk5 depletion and irradiation (8 Gy) on PARG activity in cells. No significant alteration of mean PARG activity was observed in Cdk5KD cells (p = 0.74) or irradiated Cdk5KD cells (p = 0.63) compared to controls and irradiated controls, respectively. Relative light unit values correspond to the amount of PAR polymer, which has not been hydrolyzed during a 30-min incubation with a protein extract prepared from untreated or treated cells. Data are mean ± SD of six replicates for each sample. (TIFF 213 kb)
18_2011_811_MOESM4_ESM.tif
Supplemental Fig. 4 The phenotype of Cdk5-depleted clones 1,500 and 1,501. Quantification of the relative spot intensity with time of fluorescent-tagged PARP-1 in Control or the Cdk5-depleted clone 1,500 a or clone 1,501 b and the serine-mutated tagged PARP-1 (PARP-1 MUT) in control cells. Quantitative analysis of the relative spot intensity with time of XRCC1-YFP recruitment in Control and the Cdk5-depleted clone 1,500 c or clone 1,501 d with time. Data represents mean relative spot intensity ± SEM (p < 10−4), n = 6–9 individual cells for each experimental condition. e Quantification of poly(ADP-ribose) (PAR) staining in the untreated Cdk5-depleted clones 1,500 and 1,501 and Control cells and after exposure to 8 Gy IR. Data represents the mean ± SD *** (p < 10−5) based on the fluorescence intensity in 200–300 cells/treatment group. f Western-blot analysis confirming stable depletion of Cdk5 expression in clones 1,500 and 1,501. (TIFF 2.17 mb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bolin, C., Boudra, MT., Fernet, M. et al. The impact of cyclin-dependent kinase 5 depletion on poly(ADP-ribose) polymerase activity and responses to radiation. Cell. Mol. Life Sci. 69, 951–962 (2012). https://doi.org/10.1007/s00018-011-0811-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-011-0811-6