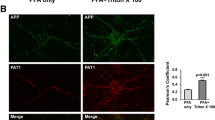
Abstract
Recently, the localization of amyloid precursor protein (APP) into reversible nanoscale supramolecular assembly or “nanodomains” has been highlighted as crucial towards understanding the onset of the molecular pathology of Alzheimer’s disease (AD). Surface expression of APP is regulated by proteins interacting with it, controlling its retention and lateral trafficking on the synaptic membrane. Here, we evaluated the involvement of a key risk factor for AD, PICALM, as a critical regulator of nanoscale dynamics of APP. Although it was enriched in the postsynaptic density, PICALM was also localized to the presynaptic active zone and the endocytic zone. PICALM colocalized with APP and formed nanodomains with distinct morphological properties in different subsynaptic regions. Next, we evaluated if this localization to subsynaptic compartments was regulated by the C-terminal sequences of APP, namely, the “Y682ENPTY687” domain. Towards this, we found that deletion of C-terminal regions of APP with partial or complete deletion of Y682ENPTY687, namely, APP–Δ9 and APP–Δ14, affected the lateral diffusion and nanoscale segregation of APP. Lateral diffusion of APP mutant APP–Δ14 sequence mimicked that of a detrimental Swedish mutant of APP, namely, APP–SWE, while APP–Δ9 diffused similar to wild-type APP. Interestingly, elevated expression of PICALM differentially altered the lateral diffusion of the APP C-terminal deletion mutants. These observations confirm that the C-terminal sequence of APP regulates its lateral diffusion and the formation of reversible nanoscale domains. Thus, when combined with autosomal dominant mutations, it generates distinct molecular patterns leading to onset of Alzheimer’s disease (AD).




Similar content being viewed by others
Availability of data and materials
Data and further details regarding the manuscript will be made available by request to the corresponding author.
References
Almeida JFF, Dos Santos LR, Trancozo M, de Paula F (2018) Updated meta-analysis of BIN1, CR1, MS4A6A, CLU, and ABCA7 variants in Alzheimer’s disease. J Mol Neurosci 64(3):471–477
Ando K, Brion JP, Stygelbout V, Suain V, Authelet M, Dedecker R, Chanut A, Lacor P, Lavaur J, Sazdovitch V, Rogaeva E, Potier MC, Duyckaerts C (2013) Clathrin adaptor CALM/PICALM is associated with neurofibrillary tangles and is cleaved in Alzheimer’s brains. Acta Neuropathol 125(6):861–878
Ando K, Nagaraj S, Kucukali F, de Fisenne MA, Kosa AC, Doeraene E, Lopez GL, Brion JP, Leroy K (2022) PICALM and Alzheimer’s disease: an update and perspectives. Cells 11(24):3994
Azarnia Tehran D, Kochlamazashvili G, Pampaloni NP, Sposini S, Shergill JK, Lehmann M, Pashkova N, Schmidt C, Lowe D, Napieczynska H, Heuser A, Plested AJR, Perrais D, Piper RC, Haucke V, Maritzen T (2022) Selective endocytosis of Ca(2+)-permeable AMPARs by the Alzheimer’s disease risk factor CALM bidirectionally controls synaptic plasticity. Sci Adv 8(21):eabl5032
Bellenguez C, Grenier-Boley B, Lambert JC (2020) Genetics of Alzheimer’s disease: where we are, and where we are going. Curr Opin Neurobiol 61:40–48
Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224(Pt 3):213–232
Borg JP, Ooi J, Levy E, Margolis B (1996) The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol 16(11):6229–6241
Chen GF, Xu TH, Yan Y, Zhou YR, Jiang Y, Melcher K, Xu HE (2017) Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol Sin 38(9):1205–1235
De Rossi P, Andrew RJ, Musial TF, Buggia-Prevot V, Xu G, Ponnusamy M, Ly H, Krause SV, Rice RC, de l’Estoile V, Valin T, Salem S, Despa F, Borchelt DR, Bindokas VP, Nicholson DA, Thinakaran G (2019) Aberrant accrual of BIN1 near Alzheimer’s disease amyloid deposits in transgenic models. Brain Pathol 29(4):485–501
Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK (1996) The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA 93(10):4804–4809
Ehrlich I, Klein M, Rumpel S, Malinow R (2007) PSD-95 is required for activity-driven synapse stabilization. Proc Natl Acad Sci USA 104(10):4176–4181
Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT (2001) Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291(5506):1051–1055
Franzmeier N, Ossenkoppele R, Brendel M, Rubinski A, Smith R, Kumar A, Mattsson-Carlgren N, Strandberg O, Duering M, Buerger K, Dichgans M, Hansson O, Ewers M (2022) Alzheimer’s Disease Neuroimaging Initiative and the Swedish Bio F.s. The BIN1 rs744373 Alzheimer’s disease risk SNP is associated with faster Abeta-associated tau accumulation and cognitive decline. Alzheimers Dement 18(1):103–115
Giannone G, Hosy E, Sibarita JB, Choquet D, Cognet L (2013) High-content super-resolution imaging of live cell by uPAINT. Methods Mol Biol 950:95–110
Guerreiro R, Bras J, Hardy J (2013) SnapShot: genetics of Alzheimer’s disease. Cell 155(4):968–968 (e961)
Harris KM, Weinberg RJ (2012) Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol 4(5):a005587
Karch CM, Goate AM (2015) Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 77(1):43–51
Kechkar A, Nair D, Heilemann M, Choquet D, Sibarita JB (2013) Real-time analysis and visualization for single-molecule based super-resolution microscopy. PLoS One 8(4):e62918
Kedia S, Mandal K, Netrakanti PR, Jose M, Sisodia SS, Nair D (2021) Nanoscale organization of Nicastrin, the substrate receptor of the gamma-secretase complex, as independent molecular domains. Mol Brain 14(1):158
Kedia S, Nair D (2020) Nanoscale rearrangement of APP organization as a therapeutic target for Alzheimer’s disease. Med Hypotheses 143:110143
Kedia S, Ramakrishna P, Netrakanti PR, Jose M, Sibarita JB, Nadkarni S, Nair D (2020) Real-time nanoscale organization of amyloid precursor protein. Nanoscale 12(15):8200–8215
Kedia S, Ramakrishna P, Netrakanti PR, Singh N, Sisodia SS, Jose M, Kumar S, Mahadevan A, Ramanan N, Nadkarni S, Nair D (2021) Alteration in synaptic nanoscale organization dictates amyloidogenic processing in Alzheimer’s disease. iScience 24(1):101924
Kedia S, Ramanan N, Nair D (2021) Quantifying molecular aggregation by super resolution microscopy within an excitatory synapse from mouse hippocampal neurons. STAR Protoc 2(2):100470
Koo EH, Squazzo SL, Selkoe DJ, Koo CH (1996) Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J Cell Sci 109(Pt 5):991–998
Kumon H, Yoshino Y, Funahashi Y, Mori H, Ueno M, Ozaki Y, Yamazaki K, Ochi S, Mori T, Iga JI, Nagai M, Nomoto M, Ueno SI (2021) PICALM mRNA expression in the blood of patients with neurodegenerative diseases and geriatric depression. J Alzheimers Dis 79(3):1055–1062
Lai A, Sisodia SS, Trowbridge IS (1995) Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J Biol Chem 270(8):3565–3573
Lichtenthaler SF, Haass C, Steiner H (2011) Regulated intramembrane proteolysis–lessons from amyloid precursor protein processing. J Neurochem 117(5):779–796
Matrone C, Luvisetto S, La Rosa LR, Tamayev R, Pignataro A, Canu N, Yang L, Barbagallo AP, Biundo F, Lombino F, Zheng H, Ammassari-Teule M, D’Adamio L (2012) Tyr682 in the Abeta-precursor protein intracellular domain regulates synaptic connectivity, cholinergic function, and cognitive performance. Aging Cell 11(6):1084–1093
Maturana-Candelas A, Gomez C, Poza J, Rodriguez-Gonzalez V, Pablo VG, Lopes AM, Pinto N, Hornero R (2021) Influence of PICALM and CLU risk variants on beta EEG activity in Alzheimer’s disease patients. Sci Rep 11(1):20465
Merthan L, Haller A, Thal DR, von Einem B, von Arnim CAF (2019) The role of PTB domain containing adaptor proteins on PICALM-mediated APP endocytosis and localization. Biochem J 476(14):2093–2109
Meyerholz A, Hinrichsen L, Groos S, Esk PC, Brandes G, Ungewickell EJ (2005) Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic 6(12):1225–1234
Miyagawa T, Ebinuma I, Morohashi Y, Hori Y, Young CM, Hattori H, Maehara T, Yokoshima S, Fukuyama T, Tsuji S, Iwatsubo T, Prendergast GC, Tomita T (2016) BIN1 regulates BACE1 intracellular trafficking and amyloid-beta production. Hum Mol Genet 25(14):2948–2958
Moreau K, Fleming A, Imarisio S, Lopez Ramirez A, Mercer JL, Jimenez-Sanchez M, Bento CF, Puri C, Zavodszky E, Siddiqi F, Lavau CP, Betton M, O’Kane CJ, Wechsler DS, Rubinsztein DC (2014) PICALM modulates autophagy activity and tau accumulation. Nat Commun 5:4998
Muller UC, Deller T, Korte M (2017) Not just amyloid: physiological functions of the amyloid precursor protein family. Nat Rev Neurosci 18(5):281–298
Nair D, Hosy E, Petersen JD, Constals A, Giannone G, Choquet D, Sibarita JB (2013) Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J Neurosci 33(32):13204–13224
Nalivaeva NN, Turner AJ (2013) The amyloid precursor protein: a biochemical enigma in brain development, function and disease. FEBS Lett 587(13):2046–2054
Narayan P, Sienski G, Bonner JM, Lin YT, Seo J, Baru V, Haque A, Milo B, Akay LA, Graziosi A, Freyzon Y, Landgraf D, Hesse WR, Valastyan J, Barrasa MI, Tsai LH, Lindquist S (2020) PICALM rescues endocytic defects caused by the Alzheimer’s disease risk factor APOE4. Cell Rep 33(1):108224
Neuner SM, Tcw J, Goate AM (2020) Genetic architecture of Alzheimer’s disease. Neurobiol Dis 143:104976
Parikh I, Fardo DW, Estus S (2014) Genetics of PICALM expression and Alzheimer’s disease. PLoS One 9(3):e91242
Philipson O, Lord A, Gumucio A, O’Callaghan P, Lannfelt L, Nilsson LN (2010) Animal models of amyloid-beta-related pathologies in Alzheimer’s disease. FEBS J 277(6):1389–1409
Pimenova AA, Raj T, Goate AM (2018) Untangling genetic risk for Alzheimer’s disease. Biol Psychiatry 83(4):300–310
Provost P (2010) Interpretation and applicability of microRNA data to the context of Alzheimer’s and age-related diseases. Aging (Albany NY) 2(3):166–169
Rajeev P, Singh N, Kechkar A, Butler C, Ramanan N, Sibarita JB, Jose M, Nair D (2022) Nanoscale regulation of Ca(2+) dependent phase transitions and real-time dynamics of SAP97/hDLG. Nat Commun 13(1):4236
Russo C, Venezia V, Repetto E, Nizzari M, Violani E, Carlo P, Schettini G (2005) The amyloid precursor protein and its network of interacting proteins: physiological and pathological implications. Brain Res Brain Res Rev 48(2):257–264
Selkoe DJ, Hardy J (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8(6):595–608
Serrano-Pozo A, Das S, Hyman BT (2021) APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol 20(1):68–80
Sheng M, Kim E (2011) The postsynaptic organization of synapses. Cold Spring Harb Perspect Biol 3(12):a005678
Sun J, Carlson-Stevermer J, Das U, Shen M, Delenclos M, Snead AM, Koo SY, Wang L, Qiao D, Loi J, Petersen AJ, Stockton M, Bhattacharyya A, Jones MV, Zhao X, McLean PJ, Sproul AA, Saha K, Roy S (2019) CRISPR/Cas9 editing of APP C-terminus attenuates beta-cleavage and promotes alpha-cleavage. Nat Commun 10(1):53
Tanwar M, Kateriya S, Nair D, Jose M (2021) Optogenetic modulation of real-time nanoscale dynamics of HCN channels using photoactivated adenylyl cyclases. RSC Chem Biol 2(3):863–875
Taylor MJ, Perrais D, Merrifield CJ (2011) A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol 9(3):e1000604
Tebar F, Bohlander SK, Sorkin A (1999) Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell 10(8):2687–2702
Thomas RS, Henson A, Gerrish A, Jones L, Williams J, Kidd EJ (2016) Decreasing the expression of PICALM reduces endocytosis and the activity of beta-secretase: implications for Alzheimer’s disease. BMC Neurosci 17(1):50
Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P (2013) Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer’s APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci USA 110(42):17071–17076
Van Acker ZP, Bretou M, Annaert W (2019) Endo-lysosomal dysregulations and late-onset Alzheimer’s disease: impact of genetic risk factors. Mol Neurodegener 14(1):20
Venkatachalapathy M, Belapurkar V, Jose M, Gautier A, Nair D (2019) Live cell super resolution imaging by radial fluctuations using fluorogen binding tags. Nanoscale 11(8):3626–3632
Voskobiynyk Y, Roth JR, Cochran JN, Rush T, Carullo NV, Mesina JS, Waqas M, Vollmer RM, Day JJ, McMahon LL, Roberson ED (2020) Alzheimer’s disease risk gene BIN1 induces Tau-dependent network hyperexcitability. Elife. https://doi.org/10.7554/eLife.57354
Wilhelm BG, Mandad S, Truckenbrodt S, Krohnert K, Schafer C, Rammner B, Koo SJ, Classen GA, Krauss M, Haucke V, Urlaub H, Rizzoli SO (2014) Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344(6187):1023–1028
Xiao Q, Gil SC, Yan P, Wang Y, Han S, Gonzales E, Perez R, Cirrito JR, Lee JM (2012) Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem 287(25):21279–21289
Yuksel M, Tacal O (2019) Trafficking and proteolytic processing of amyloid precursor protein and secretases in Alzheimer’s disease development: an up-to-date review. Eur J Pharmacol 856:172415
Zhuang L, Liu X, Shi Y, Liu X, Luo B (2019) Genetic variants of PICALM rs541458 modulate brain spontaneous activity in older adults with amnestic mild cognitive impairment. Front Neurol 10:494
Acknowledgements
The authors thank the imaging facility at the Division of Biological Sciences, IISc Bengaluru, India and IISER Pune, India. We also thank the Central Animal Facility at IISc, Bangalore, India. We thank Sandhya Subramaniam for her help with primary neuronal cultures. We thank Aishwarya Babu for contribution to protein purification and biochemical analysis. We thank Dr. Narendrakumar Ramanan for sharing resources and help with molecular strategies and technical assistance. We thank Dr. Jean-Baptiste Sibarita for helpful discussion and for sharing advanced analysis modules. We also thank Dr. David Perrais for his insightful comments on the functional molecular markers at endocytic zone.
Funding
This work was supported by generous grants from Department of Biotechnology (Innovative Biotechnologist Award to D.N., M.J.), Department of Biotechnology Genomics Engineering Taskforce (D.N.), Ramalingaswami Fellowship to D.N. and M.J.), DBT-IISc Partnership program (to D.N.), IISc-STAR program grant to D.N., CBR-FABRIC GRANT (DN), Science and Engineering Research Board (SERB-ECR award to M.J. CRG CRG/2019/006899 and CRG/2022/002726 to D.N.), Indian Institute of Science (Institute of Excellence Program) and University Grants Commission, India grants to D.N. and Tata Trusts, India program grant to D.N (Co-investigator). We also acknowledge and thank; DBT (BT/PR32489/BRB/10/1786/2019) and SERB (CRG/2019/000281) for S.R.G.S. V.B. and M.S.H.S thank UGC for JRF and SRF fellowships. S.K. thanks postdoctoral fellowship support through Department of Biotechnology, Genomics Engineering Taskforce grant awarded to D.N and N.S. thanks senior research fellowship from CSIR, India.
Author information
Authors and Affiliations
Contributions
VB and DN designed research, VB performed all the experiments unless otherwise indicated, VB and DN performed data analysis, MJ prepared neuronal cell cultures and provided reagents. MS HS and NS performed additional control experiments. VB and SK were involved generation of APP plasmids. VB, MSHS and DN wrote the manuscript. SRGS helped with design of control experiments and resources. MJ edited the manuscript. All the authors provided critical inputs for the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Ethical approval and consent to participate
Wild type Sprague–Dawley Pups were acquired from Central Animal Facility (CAF, IISc). The pups from postnatal day-0 (P0), were sacrificed to dissect the hippocampus for primary neuronal cultures. All the procedures in this study were performed according to the rules and guidelines declared in the Compendium of CPCSEA 2018 by the Committee for the Purpose of Control and Supervision of Experimental Animals (CPCSEA), Ministry of Fisheries, Animal Husbandry and Dairying, India. The research protocol (CAF/Ethics/659 and CAF/Ethics/790) was approved by the Institutional Animal Ethics Committee (IAEC) of the Indian Institute of Science.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Belapurkar, V., Mahadeva Swamy, H.S., Singh, N. et al. Real-time heterogeneity of supramolecular assembly of amyloid precursor protein is modulated by an endocytic risk factor PICALM. Cell. Mol. Life Sci. 80, 295 (2023). https://doi.org/10.1007/s00018-023-04939-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04939-w