Abstract
The genus Machaerophorus includes three perennial species (M. arequipa, M. laticarpus, and M. matthioloides) inhabiting the southern Peruvian Andes. However, based on the morphological data, this genus has been reduced for more than one hundred years to synonymy of several genera of various tribes, and its phylogenetic placement using molecular data has never been studied. Therefore, in this study we explored the phylogenetic placement of Machaerophorus to test its generic status and tribal assignment, using for this purpose ribosomal nuclear (ITS) and chloroplast (trnL-F, trnH-psbA, rps16 intron) data together with a comprehensive sampling of taxa. Results obtained here demonstrated that Machaerophorus represents a well-differentiated lineage within the South American Cremolobeae–Eudemeae–Schizopetaleae clade, and hence, the genus is reinstated and excluded from the placements in Sibara and Mathewsia. Based on molecular data and morphological affinities, we included it within the tribe Schizopetaleae. A key to Machaerophorus and the genera of Schizopetaleae is also presented.




Similar content being viewed by others
References
Al-Shehbaz IA (1989) Systematics and phylogeny of Schizopetalon (Brassicaceae). Harvard Pap Bot 1:10–46
Al-Shehbaz IA (2010) A synopsis of the genus Sibara (Brassicaceae). Harvard Pap Bot 15:139–147. https://doi.org/10.3100/025.015.0107
Al-Shehbaz IA (2012a) Machaerophorus is united with the genus Sibara (Brassicaceae). Harvard Pap Bot 17:1–2. https://doi.org/10.3100/025.017.0101
Al-Shehbaz IA (2012b) A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61:931–954
Al-Shehbaz IA (2012c) Tribe Schizopetaleae. In: Anton MA, Zuloaga FO (eds) Brassicaceae, Flora Argentina, vol. 8. Editorial Sigma, Buenos Aires, pp 202–204
Al-Shehbaz AI, Cano A, Cueva Machengo MA, Salariato DL (2018) Remarkable discoveries in the long-neglected and Peruvian-endemic genus Machaerophorus (Brassicaceae). Phytotaxa 360:114–124. https://doi.org/10.11646/phytotaxa.360.2.3
Ané C, Larget B, Baum DA, Smith SD, Rokas A (2007) Bayesian estimation of concordance among gene trees. Molec Biol Evol 24:412–426. https://doi.org/10.1093/molbev/msl170
Appel O, Al-Shehbaz IA (2003) Cruciferae. In: Kubitzki K, Bayer C (eds) The families and genera of vascular plants, vol. 5. Springer, Berlin, pp 75–174. https://doi.org/10.1007/978-3-662-07255-4_17
Baum DA (2007) Concordance trees, concordance factors, and the exploration of reticulate genealogy. Taxon 56:417–426
Chen H, Deng T, Yue J, Al-Shehbaz IA, Sun H (2016) Molecular phylogeny reveals the non-monophyly of tribe Yinshanieae (Brassicaceae) and description of a new tribe, Hillielleae. Pl Diversity 38:171–182. https://doi.org/10.1016/j.pld.2016.04.004
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest2: more models, new heuristics and parallel computing. Nat Meth 9:772. https://doi.org/10.1038/nmeth.2109
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hayek A (1911) Entwurf eines Cruciferensystems auf phylogenetischer Grundlage. Beih Bot Centralbl 27:127–335
Hooker JD (1862) Cruciferae. In: Bentham G, Hooker JD (eds) Genera plantarum, vol. 1. Reeve & Co, London, pp 57–102
Khanna KR, Rollins RC (1965) A taxonomic revision of Cremolobus (Cruciferae). Contr Gray Herb 195:135–157
Kiefer M, Schmickl R, German DA, Mandáková T, Lysak MA, Al- Shehbaz IA, Franzke A, Mummenhoff K, Stamatakis A, Koch MA (2014) BrassiBase: Introduction to a novel knowledge database on Brassicaceae evolution. Pl Cell Physiol 55:e3. https://doi.org/10.1093/pcp/pct158
Larget BR, Kotha SK, Dewey CN, Ané C (2010) BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26:2910–2911. https://doi.org/10.1093/bioinformatics/btq539
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway computing environments workshop (GCE). IEEE, New Orleans, pp. 1–8
Müller K (1868) Annales botanices systematicae [Walpers], vol. 7. Sumptibus Friderici Hofmeister, Leipzig
Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D’Amico JA, Itoua I, Strand HE, Morrison JC, Loucks CJ, Allnutt TF, Ricketts TH, Kura Y, Lamoreux JF, Wettengel WW, Hedao P, Kassem KR (2001) Terrestrial ecoregions of the world: a new map of life on Earth. Bioscience 51:933–938. https://doi.org/10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2
Prantl K (1891) Cruciferae. In: Engler A, Prantl K (eds) Die natürlichen Pflanzenfamilien III. Verlag von Wilhelm Engelmann, Leipzig, pp 145–206
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/sysbio/syy032
Rollins RC (1955) A revisionary study of the genus Menonvillea (Cruciferae). Contr Gray Herb 177:3–57
Rollins RC (1966) The genus Mathewsia (Cruciferae). Acta Bot Neerl 15:102–116. https://doi.org/10.1111/j.1438-8677.1966.tb00218.x
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Salariato DL, Al-Shehbaz IA (2014) Zuloagocardamum (Brassicaceae: Thelypodieae) a new genus of Brassicaceae from the Andes highlands of northern Argentina. Syst Bot 39:563–577. https://doi.org/10.1600/036364418X697085
Salariato DL, Zuloaga FO (2015) Taxonomic placement of Onuris hauthalii (Brassicaceae: Eudemeae), based on morphology and multilocus species tree analyses, and the recognition of the new genus Alshehbazia. Kew Bull 70:49. https://doi.org/10.1007/s12225-015-9602-9
Salariato DL, Zuloaga FO, Shehbaz IA (2013a) Molecular phylogeny of Menonvillea and recognition of the new genus Aimara (Brassicaceae: Cremolobeae). Taxon 62:1220–1234. https://doi.org/10.12705/626.6
Salariato DL, Zuloaga FO, Al-Shehbaz IA (2013b) Revision and tribal placement of the Argentinean genus Parodiodoxa (Brassicaceae). Pl Syst Evol 299:305–316. https://doi.org/10.1007/s00606-012-0722-5
Salariato DL, Zuloaga FO, Cano A, Al-Shehbaz IA (2015) Molecular phylogenetics of tribe Eudemeae (Brassicaceae) and implications for its morphology and distribution. Molec Phylogenet Evol 82:43–59. https://doi.org/10.1016/j.ympev.2014.09.030
Salariato DL, Zuloaga FO, Franzke A, Mummenhoff K, Al-Shehbaz IA (2016) Diversification patterns in the CES clade (Brassicaceae tribes Cremolobeae, Eudemeae, Schizopetaleae) in Andean South America. Bot J Linn Soc 181:543–566. https://doi.org/10.1111/boj.12430
Salariato DL, Al-Shehbaz IA, Zuloaga FO (2018) Reinstatement of the Southern Andean Genus Stenodraba (Brassicaceae) based on molecular data and insights from its environmental and geographic distribution. Syst Bot 43:35–52. https://doi.org/10.1600/036364418X697085
von Schlechtendal DFL (1857) Plantae Lechlerianae. Linnaea 28:463–542
Schulz OE (1936) Cruciferae. In: Engler A, Harms H (eds) Die natürlichen Pflanzenfamilien, vol. 17B. Verlag von Wilhelm Englemann, Leipzig, pp 227–658
Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–508. https://doi.org/10.1080/10635150290069913
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molec Biol Evol 16:1114–1116
Shimodaira H, Hasegawa M (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. https://doi.org/10.1093/bioinformatics/17.12.1246
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 57:758–771. https://doi.org/10.1080/10635150802429642
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0 beta 10. Sinauer Associates, Sunderland
Thiers B (2017) [continuously updated]. Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. Available at: http://sweetgum.nybg.org/ih. Accessed July 2018
Toro-Núñez O, Mort ME, Ruiz-Ponce E, Al-Shehbaz IA (2013) Phylogenetic relationships of Mathewsia and Schizopetalon (Brassicaceae) inferred from nrDNA and cpDNA regions: taxonomic and evolutionary insights from an Atacama Desert endemic lineage. Taxon 62:343–356. https://doi.org/10.12705/622.4
Toro-Núñez O, Al-Shehbaz IA, Mort ME (2015) Phylogenetic study with nuclear and chloroplast data and ecological niche reveals Atacama (Brassicaceae), a new monotypic genus endemic from the Andes of the Atacama Desert, Chile. Pl Syst Evol 301:1377–1396. https://doi.org/10.1007/s00606-014-1157-y
Acknowledgements
This project was funded by ANPCyT (Agencia Nacional de Promoción Científica y Tecnológica) Grant PICT-2013-1042 and CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas) Grant PIP-112-201301-00124CO. Our deep gratitude goes to Dr. Fernando O. Zuloaga for his valuable support for this work and the study of South American Brassicaceae over the years. We especially appreciate the help of the editor-in-chief Martin A. Lysak and three anonymous reviewers who provided useful suggestions that improved an earlier version of this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Martin A. Lysak.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. New sequences generated for this study. Species, specimen voucher, and Genbank accession numbers (ITS, trnL-F, trnH-psbA, rps16 intron).
Online Resource 2. Taxa and GenBank accession numbers for the ITS, trnL-F, trnH-psbA, and rps16 sequences downloaded from GenBank and used in the phylogenetic analyses.
Online Resource 3. Maximum likelihood tree for Brassicaceae generated with RAxML. a ITS dataset; b trnL-F dataset.
Online Resource 4. Features of the DNA alignments used in the phylogenetic analyses within the CES clade.
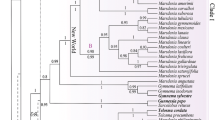
Online Resource 5. Phylogenetic placement of species of Machaerophorus within the CES clade (tribes Cremolobeae–Eudemeae–Schizopetaleae) using the individual cpDNA regions. a–c. 50% Majority rule consensus tree estimated from 6002 trees generated by Bayesian inference with MrBayes. a trnL-F dataset; b trnH-psbA dataset; c rps16 intron dataset.
Online Resource 6. Phylogenetic placement of species of Machaerophorus within the CES clade (tribes Cremolobeae–Eudemeae–Schizopetaleae). a–c. Maximum likelihood tree generated RAxML. a nrITS dataset; b. cpDNA dataset (trnL-F/trnH-psbA/rps16 intron); c concatenated nrITS + cpDNA dataset.
Online Resource 7. ITS alignment for the Brassicaceae dataset used for phylogenetic analyses in this study.
Online Resource 8. trnL-F alignment for the Brassicaceae dataset used for phylogenetic analyses in this study.
Online Resource 9. ITS alignment for the Cremolobeae–Eudemeae–Schizopetaleae (CES) dataset used for phylogenetic analyses in this study.
Online Resource 10. trnL-F alignment for the Cremolobeae–Eudemeae–Schizopetaleae (CES) dataset used for phylogenetic analyses in this study.
Online Resource 11. trnH-psbA alignment for the Cremolobeae–Eudemeae–Schizopetaleae (CES) dataset used for phylogenetic analyses in this study.
Online Resource 12. rps16 intron alignment for the Cremolobeae–Eudemeae–Schizopetaleae (CES) dataset used for phylogenetic analyses in this study.
Rights and permissions
About this article
Cite this article
Salariato, D.L., Cueva Manchego, M.A., Cano, A. et al. Phylogenetic placement of the Peruvian-endemic genus Machaerophorus (Brassicaceae) based on molecular data and implication for its systematics. Plant Syst Evol 305, 77–87 (2019). https://doi.org/10.1007/s00606-018-1553-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-018-1553-9