Abstract
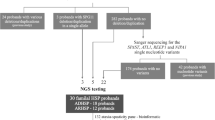
Hereditary spastic paraplegias (HSP) are a heterogeneous group of neurological disorders. Insidiously progressive spastic weakness of the lower extremities is the common criterion in all forms described. Clinically, HSP is differentiated into pure (uncomplicated) and complex (complicated) forms. While pure HSP is predominantly characterized by signs and symptoms of pyramidal tract dysfunction, additional neurological and non-neurological symptoms occur in complicated forms. Autosomal dominant, autosomal recessive, and X-linked modes of inheritance have been described and at least 48 subtypes, termed SPG1-48, have been genetically defined. Although in autosomal dominant HSP families 50–60% of etiologies can be established by genetic testing, genotype predictions based on the phenotype are limited. In order to realize high-throughput genotyping for dominant HSP, we designed a resequencing microarray for six autosomal dominant genes on the Affymetrix CustomSEQ array platform. For validation purposes, 10 previously Sanger sequenced patients with autosomal dominant HSP and 40 positive controls with known mutations in ATL1, SPAST, NIPA1, KIF5A, and BSCL2 (32 base exchanges, eight small indels) were resequenced on this array. DNA samples of 45 additional patients with AD spastic paraplegia were included in the study. With two different sequencing analysis software modules (GSEQ, SeqC), all missense/nonsense mutations in the positive controls were identified while indels had a detection rate of only 50%. In total, 244 common synonymous single-nucleotide polymorphisms (SNPs) annotated in dbSNP (build 132) corresponding to 22 distinct sequence variations were found in the 53 analyzed patients. Among the 22 different sequence variations (SPAST n = 15, ATL1 n = 3, KIF5A n = 2, HSPD1 n = 1, BSCL2 n = 1, NIPA1 n = 0), 12 were rare variants that have not been previously described and whose clinical significance is unknown. In SPAST-negative cases, a genetic diagnosis could be established in 11% by resequencing. Resequencing microarray technology can therefore efficiently be used to study genotypes and mutations in large patient cohorts.
Similar content being viewed by others
References
Fink JK, Heiman-Patterson T, Bird T, Cambi F, Dubé MP, Figlewicz DA, Fink JK, Haines JL, Heiman-Patterson T, Hentati A, Pericak-Vance MA, Raskind W, Rouleau GA, Siddique T (1996) Hereditary spastic paraplegia: advances in genetic research Hereditary Spastic Paraplegia Working group. Neurology 46(6):1507–1514
Salinas S, Proukakis C, Crosby A, Warner TT (2008) Hereditary spastic paraplegia: clinical features and pathogenic mechanisms. Lancet Neurol 7:1127–1138
Harding AE (1983) Classification of the hereditary ataxias and paraplegias. Lancet 1(8334):1151–1155
Lin P, Li J, Liu Q, Mao F, Li J, Qiu R, Hu H, Song Y, Yang Y, Gao G, Yan C, Yang W, Shao C, Gong Y (2008) A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42). Am J Hum Genet 83(6):752–759
Schlipf NA, Beetz C, Schüle R, Stevanin G, Erichsen AK, Forlani S, Zaros C, Karle K, Klebe S, Klimpe S, Durr A, Otto S, Tallaksen CM, Riess O, Brice A, Bauer P, Schöls L (2010) A total of 220 patients with autosomal dominant spastic paraplegia do not display mutations in the SLC33A1 gene (SPG42). Eur J Hum Genet 18:1065–1067
Hanein S, Martin E, Boukhris A, Byrne P, Goizet C, Hamri A, Benomar A, Lossos A, Denora P, Fernandez J, Elleuch N, Forlani S, Durr A, Feki I, Hutchinson M, Santorelli FM, Mhiri C, Brice A, Stevanin G (2008) Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am J Hum Genet 82(4):992–1002
Alazami AM, Adly N, Al Dhalaan H, Alkuraya FS (2011) A nullimorphic ERLIN2 mutation defines a complicated hereditary spastic paraplegia locus (SPG18). Neurogenetics 12(4):333–336
Dick KJ, Eckhardt M, Paisán-Ruiz C, Alshehhi AA, Proukakis C, Sibtain NA, Maier H, Sharifi R, Patton MA, Bashir W, Koul R, Raeburn S, Gieselmann V, Houlden H, Crosby AH (2010) Mutation of FA2H underlies a complicated form of hereditary spastic paraplegia (SPG35). Hum Mutat 31(4):E1251–E1260
Meilleur KG, Traoré M, Sangaré M, Britton A, Landouré G, Coulibaly S, Niaré B, Mochel F, La Pean A, Rafferty I, Watts C, Littleton-Kearney MT, Blackstone C, Singleton A, Fischbeck KH (2010) Hereditary spastic paraplegia and amyotrophy associated with a novel locus on chromosome 19. Neurogenetics 11(3):313–318
Orthmann-Murphy JL, Salsano E, Abrams CK, Bizzi A, Uziel G, Freidin MM, Lamantea E, Zeviani M, Scherer SS, Pareyson D (2009) Hereditary spastic paraplegia is a novel phenotype for GJA12/GJC2 mutations. Brain 132:426–438
Dursun U, Koroglu C, Kocasoy Orhan E, Ugur SA, Tolun A (2009) Autosomal recessive spastic paraplegia (SPG45) with mental retardation maps to 10q24.3–q25.1. Neurogenetics 10(4):325–331
Boukhris A, Feki I, Elleuch N, Miladi MI, Boland-Augé A, Truchetto J, Mundwiller E, Jezequel N, Zelenika D, Mhiri C, Brice A, Stevanin G (2010) A new locus (SPG46) maps to 9p21.2–q21.12 in a Tunisian family with a complicated autosomal recessive hereditary spastic paraplegia with mental impairment and thin corpus callosum. Neurogenetics 11(4):441–448
Blumkin L, Lerman-Sagie T, Lev D, Yosovich K, Leshinsky-Silver E (2011) A new locus (SPG47) maps to 1p13.2–1p12 in an Arabic family with complicated autosomal recessive hereditary spastic paraplegia and thin corpus callosum. J Neurol Sci 305(1–2):67–70, Epub 2011 Mar 25
Słabicki M, Theis M, Krastev DB, Samsonov S, Mundwiller E, Junqueira M, Paszkowski-Rogacz M, Teyra J, Heninger AK, Poser I, Prieur F, Truchetto J, Confavreux C, Marelli C, Durr A, Camdessanche JP, Brice A, Shevchenko A, Pisabarro MT, Stevanin G, Buchholz F (2010) A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol 8(6):e1000408
Erichsen AK, Koht J, Stray-Pedersen A, Abdelnoor M, Tallaksen CM (2009) Prevalence of hereditary ataxia and spastic paraplegia in southeast Norway: a population-based study. Brain 132(Pt 6):1577–1588
Braschinsky M, Luus SM, Gross-Paju K, Haldre S (2009) The prevalence of hereditary spastic paraplegia and the occurrence of SPG4 mutations in Estonia. Neuroepidemiology 32(2):89–93
McMonagle P, Webb S, Hutchinson M (2002) The prevalence of “pure” autosomal dominant hereditary spastic paraparesis in the island of Ireland. J Neurol Neurosurg Psychiatry 72(1):43–46
Silva MC, Coutinho P, Pinheiro CD, Neves JM, Serrano P (1997) Hereditary ataxias and spastic paraplegias: methodological aspects of a prevalence study in Portugal. J Clin Epidemiol 50(12):1377–1384
Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Dürr A, Wincker P, Brottier P, Cattolico L, Barbe V, Burgunder JM, Prud’homme JF, Brice A, Fontaine B, Heilig B, Weissenbach J (1999) Spastin, a new AAA protein is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303
Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN (2009) The human gene mutation database: 2008 update. Genome Med 1(1):13
Ribaï P, Depienne C, Fedirko E, Jothy AC, Viveweger C, Hahn-Barma V, Brice A, Durr A (2008) Mental deficiency in three families with SPG4 spastic paraplegia. Eur J Hum Genet 16(1):97–104, ai 2008
Svenstrup K, Bross P, Koefoed P, Hjermind LE, Eiberg H, Born AP, Vissing J, Gyllenborg J, Nørremølle A, Hasholt L, Nielsen JE (2009) Sequence variants in SPAST, SPG3A and HSPD1 in hereditary spastic paraplegia. J Neurol Sci 284(1–2):90–95
Battini R, Fogli A, Borghetti D, Michelucci A, Perazza S, Baldinotti F, Conidi ME, Ferreri MI, Simi P, Cioni G (2011) Clinical and genetic findings in a series of Italian children with pure hereditary spastic paraplegia. Eur J Neurol 18(1):150–157
de Bot ST, van den Elzen RT, Mensenkamp AR, Schelhaas HJ, Willemsen MA, Knoers NV, Kremer HP, van de Warrenburg BP, Scheffer H (2010) Hereditary spastic paraplegia due to SPAST mutations in 151 Dutch patients: new clinical aspects and 27 novel mutations. J Neurol Neurosurg Psychiatry 81(10):1073–1078
Alvarez V, Sánchez-Ferrero E, Beetz C, Díaz M, Alonso B, Corao AI, Gámez J, Esteban J, Gonzalo JF, Pascual-Pascual SI, López de Munain A, Moris G, Ribacoba R, Márquez C, Rosell J, Marín R, García-Barcina MJ, Del Castillo E, Benito C, Coto E (2010) Mutational spectrum of the SPG4 (SPAST) and SPG3A (ATL1) genes in Spanish patients with hereditary spastic paraplegia. BMC Neurol 10:89
Shoukier M, Neesen J, Sauter SM, Argyriou L, Doerwald N, Pantakani DV, Mannan AU (2009) Expansion of mutation spectrum, determination of mutation cluster regions and predictive structural classification of SPAST mutations in hereditary spastic paraplegia. Eur J Hum Genet 17(2):187–194, Erratum in: Eur J Hum Genet. 2009 Mar;17(3):401-2
McCorquodale Iii D, Ozomaro U, Huang J, Montenegro G, Kushman A, Citrigno L, Price J, Speziani F, Pericak-Vance MA, Züchner S. Mutation screening of spastin, atlastin, and REEP1 in hereditary spastic paraplegia. Clin Genet. 2010 Jul 2. [Epub ahead of print]
Tamm R (2010) Novel human pathological mutations. Gene symbol: SPAST. Disease: hereditary spastic paraplegia. Hum Genet 127(1):112
Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, Apak M, Heiman-Patterson T, Ming L, Bui M, Fink JK (2001) Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet 29:326–331
Züchner S, Wang G, Tran-Viet KN, Nance MA, Gaskell PC, Vance JM, Ashley-Koch AE, Pericak-Vance MA (2006) Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet 79:365–369
Beetz C, Schüle R, Deconinck T, Tran-Viet KN, Zhu H, Kremer BP, Frints SG, van Zelst-Stams WA, Byrne P, Otto S, Nygren AO, Baets J, Smets K, Ceulemans B, Dan B, Nagan N, Kassubek J, Klimpe S, Klopstock T, Stolze H, Smeets HJ, Schrander-Stumpel CT, Hutchinson M, van de Warrenburg BP, Braastad C, Deufel T, Pericak-Vance M, Schöls L, de Jonghe P, Züchner S (2008) REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain 131(Pt 4):1078–1086
Valdmanis PN, Meijer IA, Reynolds A, Lei A, MacLeod P, Schlesinger D, Zatz M, Reid E, Dion PA, Drapeau P, Rouleau GA (2007) Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet 80:152–161
Rocco P, Vainzof M, Froehner SC, Peters MF, Marie SK, Passos-Bueno MR, Zatz M (2000) Brazilian family with pure autosomal dominant spastic paraplegia maps to 8q: analysis of muscle beta 1 syntrophin. Am J Med Genet 92(2):122–127
Schroeder C, Stutzmann F, Weber BH, Riess O, Bonin M (2010) High-throughput resequencing in the diagnosis of BRCA1/2 mutations using oligonucleotide resequencing microarrays. Breast Cancer Res Treat 122:287–297
Denning L, Anderson JA, Davis R, Gregg JP, Kuzdenyi J, Maselli RA (2007) High throughput genetic analysis of congenital myastenic syndromes using resequencing microarrays. PLoS One 2(9):e918
Waldmüller S, Müller M, Rackebrandt K, Binner P, Poths S, Bonin M, Scheffold T (2008) Array-based resequencing assay for mutations causing hypertrophic cardiomyopathy. Clinical Chemistry 54(4):682–687
Kothiyal P, Cox S, Ebert J, Husami A, Kenna MA, Greinwald JH, Aronow BJ, Rehm HL (2010) High-throughput detection of mutations responsible for childhood hearing loss using resequencing microarrays. BMC Biotechnology 10:10
Hacia JG (1999) Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet 21:42–47
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12(6):996–1006, http://genome.ucsc.edu/
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Bachmann HS, Siffert W, Frey UH (2003) Successful amplification of extremely GC-rich promoter regions using a novel ‘slowdown PCR’ technique. Pharmacogenetics 13(12):759–766
Beetz C, Schüle R, Klebe S, Klimpe S, Klopstock T, Lacour A, Otto S, Sperfeld AD, van de Warrenburg B, Schöls L, Deufel T (2008) Screening of hereditary spastic paraplegia patients for alterations at NIPA1 mutational hotspots. J Neurol Sci 268(1–2):131–135
Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C, Mathews DJ, Shah NA, Eichler EE, Warrington JA, Chakravarti A (2001) High-throughput variation detection and genotyping using microarrays. Genome Res 11(11):1913–1925
Schwarz A, Roedelsperger A, Schnelke A, Seelow A (2010) MutationTaster-evaluates disease-causing potential of sequence alterations. Nature Methods 7:575–576
Mandal NA, Heckenlively JR, Burch T, Chen L, Vasireddy V, Koenekoop RK, Sieving PA, Ayyagari R (2005) Sequencing arrays for screening multiple genes associated with early-onset human retinal degradations on a high-throughput platform. Invest Ophthalmol Vis Sci 46(9):3355–3362
Xu N, Podolsky RH, Chudgar P, Chorich LP, Liu C, McDonough PG, Warrington JA, Layman LC (2005) Screening candidate genes for mutations in patients with hypogonadism using custom genome resequencing microarrays. Am J Obstet Gynecol 192:1274–1284
Schlipf N, Schüle R, Klimpe S, Karle K, Synofzik M, Schicks J, Riess O, Schöls L, Bauer P (2011) Amplicon-based high-throughput pooled sequencing identifies mutations in CYP7B1 and SPG7 in sporadic spastic paraplegia patients. Clin Genet 80(2):148–160
Beetz C, Nygren AO, Schickel J, Auer-Grumbach M, Bürk K, Heide G, Kassubek J, Klimpe S, Klopstock T, Kreuz F, Otto S, Schüle R, Schöls L, Sperfeld AD, Witte OW, Deufel T (2006) High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology 67(11):1926–1930
Svenson IK, Ashley-Koch AE, Pericak-Vance MA, Marchuk DA (2001) Identification and expression analysis of spastin gene mutations in hereditary spastic paraplegia. Am J Hum Genet 68(5):1077–1085
Sauter S, Miterski B, Klimpe S, Bönsch D, Schöls L, Visbeck A, Papke T, Hopf HC, Engel W, Deufel T, Epplen JT, Neesen J (2002) Mutation analysis of the spastin gene (SPG4) in patients in Germany with autosomal dominant hereditary spastic paraplegia. Hum Mutat 20(2):127–132
Patrono C, Scarano V, Cricchi F, Melone MA, Chiriaco M, Napolitano A, Malandrini A, De Michele G, Petrozzi L, Giraldi C, Santoro L, Servidei S, Casali C, Filla A, Santorelli FM (2005) Autosomal dominant hereditary spastic paraplegia: DHPLC-based mutation analysis of SPG4 reveals eleven novel mutations. Hum Mutat 25(5):506
Lindsey JC, Lusher ME, McDermott CJ, White KD, Reid E, Rubinsztein DC, Bashir R, Hazan J, Shaw PJ, Bushby KM (2000) Mutation analysis of the spastin gene (SPG4) in patients with hereditary spastic paraparesis. J Med Genet 37(10):759–765
Fonknechten N, Mavel D, Byrne P, Davoine CS, Cruaud C, Bönsch D, Samson D, Coutinho P, Hutchinson M, McMonagle P, Burgunder JM, Tartaglione A, Heinzlef O, Feki I, Deufel T, Parfrey N, Brice A, Fontaine B, Prud’homme JF, Weissenbach J, Dürr A, Hazan J (2000) Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum Mol Genet 9(4):637–644
Montenegro G, Rebelo AP, Connell J, Allison R, Babalini C, D’Aloia M, Montieri P, Schüle R, Ishiura H, Price J, Strickland A, Gonzalez MA, Baumbach-Reardon L, Deconinck T, Huang J, Bernardi G, Vance JM, Rogers MT, Tsuji S, De Jonghe P, Pericak-Vance MA, Schöls L, Orlacchio A, Reid E, Züchner S (2012) Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J Clin Invest. doi:10.1172/JCI60560 [Epub ahead of print]
Huppke P, Brendel C, Kalscheuer V, Korenke GC, Marquardt I, Freisinger P, Christodoulou J, Hillebrand M, Pitelet G, Wilson C, Gruber-Sedlmayr U, Ullmann R, Haas S, Elpeleg O, Nürnberg G, Nürnberg P, Dad S, Møller LB, Kaler SG, Gärtner J (2012) Mutations in SLC33A1 cause a lethal autosomal-recessive disorder with congenital cataracts, hearing loss, and low serum copper and ceruloplasmin. Am J Hum Genet 90(1):61–68
De Leeneer K, Hellemans J, De Schrijver J, Baetens M, Poppe B, Van Criekinge W, De Paepe A, Coucke P, Claes K (2011) Massive parallel amplicon sequencing of the breast cancer genes BRCA1 and BRCA2 opportunities, challenges, and limitations. Hum Mutat 32(3):335–344
Acknowledgments
We thank the German Network of Hereditary Movement Disorders (GeNe Move) and the Agence National pour la Recherche for funding support, as well as Anna Sowa for language editing.
Ethical standards
The authors declare that the experiments comply with current laws in Germany.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Claudia Dufke and Nina Schlipf contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
(XLS 22 kb)
Online Resource 2
(XLS 51 kb)
Online Resource 3
(XLS 31 kb)
Rights and permissions
About this article
Cite this article
Dufke, C., Schlipf, N., Schüle, R. et al. A high-throughput resequencing microarray for autosomal dominant spastic paraplegia genes. Neurogenetics 13, 215–227 (2012). https://doi.org/10.1007/s10048-012-0329-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-012-0329-6