Abstract
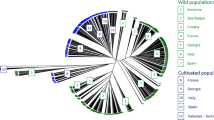
Grapevine (Vitis vinifera L.) is one of the oldest and most important fruit crops in the world. Iran is considered as one of the regions where grapevine plants have been used and taken into cultivation. In the present study, 12 simple sequence repeat (SSR) markers were used to evaluate the genetic diversity among Iranian grape cultivars and their relationships with Italian ones. The observed number of alleles (N) per locus varied from 6 to 20 and also the number of effective alleles (Ne) ranged from 1.34 to 2.00 among cultivars, indicating that the SSRs were highly informative. The polymorphism information content (PIC) values ranged from 0.49 to 0.87 and classified the six loci as highly informative markers (PIC > 0.70). The mean observed heterozygosity (Ho) was higher (0.85) than the mean expected heterozygosity (He) (0.43), demonstrating a random union of gametes in the population. Genetic similarity among cultivars ranged from 0.14 to 0.93, indicating high genetic variation among them. UPGMA and Bayesian clustering analyses revealed high genetic variation among studied cultivars and grouped them into two main clusters. The present findings might render striking information in breeding management strategies for genetic conservation and cultivar improvement.



Similar content being viewed by others
References
Adam-Blondon AF, Roux C, Claux D, Butterlin G, Merdinoglu D, This P (2004) Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet 109:1017–1027
Agar G, Yildirim N, Ercisli S, Ergul l, Yuksel C (2012) Determination of genetic diversity of Vitis vinifera cv. Kabarcik populations from the Coruh Valley using SSR markers. Biochem Genet 50:476–483
Arnold C, Gillet F, Cobat JM (1998) Situation de la vigne sauvage (Vitis vinifera subsp. sylvestris) en Europe. Vitis 37:159–170
Bandelj D, Jaksˇe J, Javornik B (2004) Assessment of genetic variability of olive varieties by microsatellite and AFLP markers. Euphytica 136:93–102
Beridze T, Pipia I, Beck J, Hsu SC, Gamkrelidze M, Gognashvili M, Tabidze V, This P, Bacilieri R, Gotsiridze V, Glonti M, Schaal B (2011) Plastid DNA sequence diversity in a worldwide set of grapevine cultivars (Vitis vinifera L. subsp. sativa). Bull Georgian Natl Acad Sci 5:98–103
Bikash D, Ranvir S, Kumar S (2008) Fruit based agro-forestry system for uplands. Horticulture and AgroForestry Research Programme. ICAR Research Complex for Eastern Region Ranchi, Jharkhand, India, pp 341–345
Borrego J, Rodriguez I, Andres MT, Martin J, Chavez J, Cabello F, Ibanez J (2001) Characterization of the most important Spanish grape varieties through isoenzyme and microsatellite analysis. Proc Int Symp Mol Markers Acta Hortic 546:371–375
Boz Y, Bakir M, Celikkol BP (2011) Genetic characterization of grape (Vitis vinifera L.) germplasm from Southeast Anatolia by SSR markers. Vitis. 50(3):99–106
Buteler MI, Jarret RL, LaBonte DR (1999) Sequence characterization of microsatellites in diploid and polyploid Ipomea. Theor Appl Genet 99:123–132
Commbe BG (1992) Research on development and ripening of the grape berry. Am J Enol Viticult 43:101–110
Crespan M (2003) The parentage of Muscat of Hamburg. Vitis 42:193–197
Crespan M, Milani N (2001) The muscats: a molecular analysis of synonyms, homonyms and genetic relationships within a large family of grapevine cultivars. Vitis 40:23–30
Dalbo MA, Ye GN, Weeden NF, Steinkellner H, Sefc KM, Reisch BI (2000) A gene controlling sex in grapevine placed on a molecular marker based genetic map. Genome 43:333–340
Doliguez A, Bouquet A, Danglot Y, Lahogue F, Riaz S, Meredith CP, Edwards KJ, This P (2002) Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor Appl Genet 105:780–795
Doulati-Baneh H, Mohammadi SA, Labra M (2013) Genetic structure and diversity analysis in Vitis vinifera L. cultivars from Iran using SSR markers. Sci Hortic 160:29–36
Dzhambazova T, Tsvetkov I, Atanassov I (2009) Genetic diversity in native Bulgarian grapevine germplasm (Vitis vinifera L.) based on nuclear and chloroplast microsatellite polymorphisms. Vitis. 48(3):115–121
Ekhvaia J, Gurushidze M, Blattner FR, Akhalkatsi M (2014) Genetic diversity of Vitis vinifera in Georgia: relationships between local cultivars and wild grapevine, V. vinifera L. subsp. sylvestris. Genet Resour Crop Evol 61:1507–1521
Ergul A, Kazan K, Aras S, Cevik V, Celik H, Soylemezoglu G (2006) AFLP analysis of genetic variation within the two economically important Anatolian grapevine (Vitis vinifera L.) varietal groups. Genome 49:467–495
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Eyduran SP, Ercisli S, Akin M, Eyduran E (2016) Genetic characterization of autochthonous grapevine cultivars from Eastern Turkey by simple sequence repeats (SSRs). Biotechnol Biotechnol Equip 30:26–31
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Frank T, Botta R, Thomas MR (2002) Chimerism in grapevine: implications for cultivar identity, ancestry and genetic improvement. Theor Appl Genet 204:192–199
Gago P, Santiago JL, Boso S, Alonso-Villaverde V, Grando S, Martinez C (2009) Biodiversity and characterization of twenty-two Vitis vinifera L. cultivars in the Northwestern Iberian Peninsula. Am J Enol Viticult 60:293–301
GENRES-081 (2001) Primary and secondary descriptors list for grapevine cultivars and species (Vitis L.) Institut für Rebenzüchtung Geilweilerhof, Siebeldingen. http://www.genres.de/vitis/
Gheorghe RN, Popescu CF, Pamfil D (2010) Genetic diversity of some Romanian grapevine cultivars as revealed by microsatellite markers. Rom Biotechnol Lett 15(2):26–31
Gismondi A, Canini A (2013) Microsatellite analysis of Latial Olea europaea L. cultivars. Plant Biosyst 147(3):686–691
Gismondi A, Fanali F, Martìnez-Labarga JM, Grilli Caiola M, Canini A (2013) Crocus sativus L. genomics and different DNA barcode applications. Plant Syst Evol 299(10):1859–1863
Gismondi A, Impei S, Di Marco G, Crespan M, Leonardi D, Canini A (2014) Detection of new genetic profiles and allelic variants in improperly classified grapevine accessions. Genome 57:111–118
Godarzian P, Erfanifard SY, Sadeghi H (2013) Detection and classification of available agroforestry systems in Fars province (case study: Kazerun city). Sus Agric Prod Sci 23:56–70
Goudarzian P, Yazdani MR (2015) Climate diversity in line with agroforestry systems: studying technicalities of agroforestry systems and allied components in two diverse climatic regions (Warm climate vs. cold climate) (Case study: Kazeroun and Sepidan in Fars Province, I.R.Iran). Desert 20:157–166
Grassi F, Labra M, Imazio S, Ocete R, Failla O, Scienza A, Sala F (2006) Phylogeographical structure and conservation genetics of wild grapevine. Conserv Genet 7:837–845
Haidari M, Lotfalian M, Tashakori M, Valipour A (2016) Investigating the local utilization of forest in north Zagros (Case study: Baneh Region) Iranian Journal of. Forest 8:313–331
Hocquigny S, Pelsy F, Dumas V, Kindt S, Heloir MC, Merdinoglu D (2004) Diversification within grapevine cultivars goes through chimeric states. Genome 47:579–589
Huglin P (1988) Biologie et e`cologie de la vigne, 2nd edn. Tec and Doc, Bordeaux
Hvarleva T, Rusanov K, Lefort F et al (2004) Genotyping of Bulgarian Vitis vinifera L. cultivars by microsatellite analysis. Vitis. 43:27–34
Ibanez J, Andres MT, Molino A, Borrego J (2003) Genetic study of key Spanish grapevine varieties using microsatellite analysis. Am J Enol Viticult 54(1):22–30
Imazio S, Maghradze D, De Lorenzis G, Bacilieri R, Laucou V, This P, Scienza A, Kaill O (2013) From the cradle of grapevine domestication: molecular overview and description of Georgian grapevine (Vitis vinifera L.) germplasm. Tree Genet Genomes 9:641–658
Khadivi-Khub A, Salimpour A, Rasouli M (2014) Analysis of grape germplasm from Iran based on fruit characteristics. Braz J Bot 37:105–113
Lefort F, Roubelakis-Angelakis KA (2001) Genetic comparison of Greek cultivars of Vitis vinifera L. by nuclear microsatellite profiling. Am J Ecol Vitic 52(2):101–108
Lopes MS, Sefc KM, Dias EE, Steinkellner H, Machado MLD, Machado AD (1999) The use of microsatellites for germplasm management in a Portuguese grapevine collection. Theor Appl Genet 99(3–4):733–739
Lopes MS, Mendonca D, Rodrigues dos Santos M, Eiras-Dias JE, da Camara Machado A (2009) New insights on the genetic basis of Portuguese grapevine and on grapevine domestication. Genome 52:790–800
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Martin JP, Borrego J, Cabello F, Ortiz JM (2003) Characterization of Spanish grapevine cultivar diversity using sequence-tagged microsatellite site markers. Genome 46:10–18
Martin JP, Santiago PC, Leel F, Martinez MC, Ortiz JM (2006) Determination of relationships among autochthonous grapevine varieties (Vitis vinifera L.) in the Northwest of the Iberian Peninsula by using microsatellite markers. Genet Resour Crop Evol 53:1255–1261
Matinkhah SH, Shamekhi T, Khajedin G, Jafari M, Jalalian A (2003) Developing a method for diagnosis and characterization of traditional agroforestry systems in Iran (Case Study: Kohkiloieh and Boyerahmad Province). Iran J Natural Res 56:213–229
McGovern PE (2003) Ancient wine. Princeton University Press, Princeton
Mcgovern PE, Glusker DL, Exner LJ, Voigt MM (1996) Neolithic resinated wine. Nature 381:480–481
Moreno-Sanz P, Dolores Loureiro M, Suárez B (2011) Microsatellite characterization of grapevine (Vitis vinifera L.) genetic diversity in Asturias (Northern Spain). Sci Hortic 129:433–440
Nunez Y, Fresno J, Torres V, Ponz F, Gallego FJ (2004) Practical use of microsatellite markers to manage Vitis vinifera germplasm: molecular identification of grapevine samples collected blindly in D.O. ‘‘El Bierzo’’ (Spain). J Hortic Sci Biotech 79(3):437–440
Olmo HP (1976) Grapes. In: Simmonds NW (ed) Evolution of crop plants. Longman, London, pp 294–298
PapannA N, Rao V, Murthy S, Simon L (2009) Microsatellite-based genetic diversity assessment in grape (Vitis vinifera L.) germplasm and its relationship with agronomic traits. Int J Fruit Sci 9:92–105
Pavlicek A, Hrda S, Flegr J (1999) FreeTree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Folia Biol 45:97–99
Peakall R, Smouse PE (2006) genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Pipia I, Gogniashvili M, Tabidze V, Beridze T, Gamkrelidze M, Gotsiridze V, Melyan G, Musayev M, Salimov V, Beck J, Schaal B (2012) Plastid DNA sequence diversity in wild grapevine samples (Vitis vinifera subsp. sylvestris) from the Caucasus region. Vitis 51:119–124
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Riaz S, Garrison KE, Dangl GS, Boursiquot JM, Meredith CP (2002) Genetic divergence and chimerism within ancient asexually propagated winegrape cultivars. J Am Soc Hort Sci 127:508–514
Riaz S, Dangl GS, Edwards KJ, Meredith CP (2004) A microsatellite marker based framework map of Vitis vinifera L. Theor Appl Genet 108:864–872
Rohlf FJ (2000) NTSYS-pc numerical taxonomy and multivariate analysis system. Version 2.1. Exeter Software, Setauket
Romesburg HC (1990) Cluster analysis for researchers. Krieger Publishing, Malabar
Santana JC, Hidalgo E, De Lucas AU, Recio P, Ortiz JM, Martin JP, Yuste J, Arranz C, Rubio JA (2008) Identification and relationships of accessions grown in the grapevine (Vitis vinifera L.) Germplasm Bank of Castilla y Leon (Spain) and the varieties authorized in the VQPRD areas of the region by SSR-marker analysis. Genet Resour Crop Evol 55:573–583
Sefc KM, Lefort F, Gando MS, Scott KD, Steinkeller H, Thomas MR (2001) Microsatellite markers for grapevine: a state of the art. In: Roubelakis-Angelakis KA (ed) Molecular biology and biotechnology of the grapevine. Kluwer Academic Publishers, Amsterdam, pp 433–463
Sefc KM, Steinkellner H, Lefort F, Botta R, Da Camara Machado A, Borrego J, Maletic E, Glo¨ssl H (2003) Evaluation of the genetic contribution of local wild vines to European grapevine cultivars. Am J Enol Viticul 54:15–21
Selli F, Bakir M, Inan G, Aygun H, Boz Y, Yasasin AS, Ozer C, Akman B, Soylemezoglu G, Kazan K, Ergul A (2007) Simple sequence repeat-based assessment of genetic diversity in ‘Dimrit’ and ‘Germe’ grapevine accessions from Turkey. Vitis 46:182–187
Shahabian M, Mostashari M, Malakouti MJ (2001) Effect of different methods of fertilisation on grape yield in Qazvin. In: Horst WJ (ed) Plant Nutrition. Developments in Plant and Soil Sciences, vol 92. Springer, Dordrecht, pp 790–791
Snoussi H, Harbi Ben Slimane M, Ruiz-Garcia L, Zapataer JM, Arroyo-Garcia R (2004) Genetic relationships among cultivated and wild grapevine accessions from Tunisia. Genome 47:1211–1219
Stajner N, Korosec-Koruza Rusjan D (2008) Microsatellite genotyping of old Slovenian grapevine varieties (Vitis vinifera L.) of the Primorje (coastal) winegrowing region. Vitis 47(4):201–204
Stajner N, Rusjan D, Korosec-Koruza Z (2011) Genetic characterization of old Slovenian grapevine varieties of Vitis vinifera L. by microsatellite genotyping. Am J Enol Vitic 62(2):250–255
Stajner N, Tomic L, Ivanisevic D (2014) Microsatellite inferred genetic diversity and structure of Western Balkan grapevines (Vitis vinifera L.). Tree Genet Genomes 10:127–140
Tangolar SG, Soydam S, Bakir M, Karaagac E, Tangolar S, Ergul A (2009) Genetic analysis of grapevine cultivars from the eastern Mediterranean region of Turkey, based on SSR markers. Tarim Bilim Derg 15(1):1–8
This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L, Maul E (2004) Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet 109(7):1448–1458
This P, Lacombe T, Tomas M (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22:511–519
Tomic L, Stajner N, Jovanovic-Cvetkovic T (2012) Identity and genetic relatedness of Bosnia and Herzegovina grapevine germplasm. Sci Hortic 143:122–126
Vafaee Y, Ghaderi N, Khadivi A (2017) Morphological variation and marker-fruit trait associations in a collection of grape (Vitis vinifera L.). Sci Hortic 225:771–782
Vavilov NI (1931) Dikie rodichi plodovikh dereviev Aziatskoi chasti SSSR I Kvakaza I problema proiskhozhdenia plodovikh dereveiev (Wild progenitors of the fruit trees of Turkestan and the Caucasus and the problem of the origin of the fruit trees). Bull App Bot Genet Plant Breed 26:85–134 (in Russian)
Vignani R, Scali M, Masi E, Cresti M (2002) Genomic variability in Vitis vinifera L. ‘Sangiovese’ assessed by microsatellite and non-radioactive AFLP test. Electron J Biotech 5:1–11
White E, Sahota R, Edes S (2002) Rapid microsatellite analysis using discontinuous polyacrylamide gel electrophoresis. Genome 45:1107–1109
Zecca G, Grassi F (2013) RPB2 gene reveals phylodemographic signal in wild and domesticated grapevine (Vitis vinifera). J Syst Evol 51:205–211
Zohary D, Hopf M (2000) Domestication of plants in the Old World. University Press, Oxford
Zulini L, Russo M, Peterlunger E (2002) Genotyping wine and table grape cultivars from Apulia (Southern Italy) using microsatellite markers. Vitis 41:183–187
Acknowledgements
The authors would like to thank Arak University in Iran and University of Rome ‘‘Tor Vergata’’ in Italy for funding this research work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khadivi, A., Gismondi, A. & Canini, A. Genetic characterization of Iranian grapes (Vitis vinifera L.) and their relationships with Italian ecotypes. Agroforest Syst 93, 435–447 (2019). https://doi.org/10.1007/s10457-017-0134-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10457-017-0134-1