Abstract
Terminology extraction is an essential task in domain knowledge acquisition, as well as for information retrieval. It is also a mandatory first step aimed at building/enriching terminologies and ontologies. As often proposed in the literature, existing terminology extraction methods feature linguistic and statistical aspects and solve some problems related (but not completely) to term extraction, e.g. noise, silence, low frequency, large-corpora, complexity of the multi-word term extraction process. In contrast, we propose a cutting edge methodology to extract and to rank biomedical terms, covering all the mentioned problems. This methodology offers several measures based on linguistic, statistical, graphic and web aspects. These measures extract and rank candidate terms with excellent precision: we demonstrate that they outperform previously reported precision results for automatic term extraction, and work with different languages (English, French, and Spanish). We also demonstrate how the use of graphs and the web to assess the significance of a term candidate, enables us to outperform precision results. We evaluated our methodology on the biomedical GENIA and LabTestsOnline corpora and compared it with previously reported measures.
Similar content being viewed by others
1 Introduction
The huge amount of biomedical data available today often consists of plain text fields, e.g. clinical trial descriptions, adverse event reports, electronic health records, emails or notes expressed by patients within forums (Murdoch and Detsky 2013). These texts are often written using a specific language (expressions and terms) of the associated community. Therefore, there is a need for formalization and cataloging of these technical terms or concepts via the construction of terminologies and ontologies (Rubin et al. 2008). These technical terms are also important for information retrieval (IR), for instance when indexing documents or formulating queries. However, as the task of manually extracting terms of a domain is very long and cumbersome, researchers have striving to design automatic methods to assist knowledge experts in the process of cataloging the terms and concepts of a domain under the form of vocabularies, thesauri, terminologies or ontologies.
Automatic term extraction (ATE), or automatic term recognition (ATR), is a domain which aims to automatically extract technical terminology from a given text corpus. We define technical terminology as the set of terms used in a domain. Term extraction is an essential task in domain knowledge acquisition because the technical terminology can be used for lexicon updating, domain ontology construction, summarization, named entity recognition or, as previously mentioned, IR.
In the biomedical domain, there is a substantial difference between existing resources (hereafter called terminologies or ontologies) in English, French, and Spanish. In English, there are about 9,919,000 terms associated with about 8,864,000 concepts such as those in UMLSFootnote 1 or BioPortal (Noy et al. 2009). Whereas in French there are only about 330,000 terms associated with about 160,000 concepts (Névéol et al. 2014), and in Spanish 1,172,000 terms associated with about 1,140,000 concepts. Note the strong difference in the number of ontologies and terminologies available in French or Spanish. This makes ATE even more important for these languages.
In biomedical ontologies, different terms may be linked to the same concept and are semantically similar with different writing, for instance “neoplasm” and “cancer” in MeSH or SNOMED-CT. Ontologies also contain terms with morphosyntaxic variants, for instance plurals like ‘‘external fistula” and “external fistulas”, and this group of variants is linked to a preferred term. As one of our goals is to extract new terms to enrich ontologies, our approach does not normalize variant terms, mainly because normalization would lead to penalization in extracting new variant terms. Technical terms are useful to gain further insight into the conceptual structure of a domain. These may be: (i) single-word terms (simple), or (ii) multi-word terms (complex). The proposed study focuses on both cases.
Term extraction methods usually involve two main steps. The first step extracts candidate terms by unithood calculation to qualify a string as a valid term, while the second step verifies them through termhood measures to validate their domain specificity. Formally, unithood refers to the degree of strength or stability of syntagmatic combinations and collocations, and termhood is defined as the degree to which a linguistic unit is related to domain-specific concepts (Kageura and Umino 1996). ATE has been applied to several domains, e.g. biomedical (Lossio-Ventura et al. 2014c; Frantzi et al. 2000; Zhang et al. 2008; Newman et al. 2012), ecological (Conrado et al. 2013), mathematical (Stoykova and Petkova 2012), social networks (Lossio-Ventura et al. 2012), banking (Dobrov and Loukachevitch 2011), natural sciences (Dobrov and Loukachevitch 2011), information technology (Newman et al. 2012; Yang et al. 2009), legal (Yang et al. 2009), as well as post-graduate school websites (Qureshi et al. 2012).
The main issues in ATE are: (i) extraction of non-valid terms (noise) or omission of terms with low frequency (silence), (ii) extraction of multi-word terms having various complex various structures, (iii) manual validation efforts of the candidate terms (Conrado et al. 2013), and (iv) management of large-scale corpora. Inspired by our previously published results and in response to the above issues, we propose a cutting edge methodology to extract biomedical terms. We propose new measures and some modifications of existing baseline measures. Those measures are divided into: (1) ranking measures, and (2) re-ranking measures. Our ranking measures are statistical- and linguistic-based and address issues (i), (ii) and (iv). Our two re-ranking measures the first one called TeRGraph is a graph-based measure which deals with issues (i), (ii) and (iii). The second one, called WAHI, is a web-based measure which also deals with issues (i), (ii) and (iii). The novelty of the WAHI measure is that it is web-based which has, to the best of our knowledge, never been applied within ATE approaches.
The main contributions of our article are: (1) enhanced consideration of the term unithood, by computing a degree of quality for the term unithood, and, (2) consideration of the term dependence in the ATE process. The quality of the proposed methodology is highlighted by comparing the results obtained with the most commonly used baseline measures. Our evaluation experiments were conducted despite difficulties in comparing ATE measures, mainly because of the size of the corpora used and the lack of available libraries associated with previous studies. Our three measures improve the process of automatic extraction of domain-specific terms from text collections that do not offer reliable statistical evidence (i.e. low frequency).
The paper is organized as follows. We first discuss related work in Sect. 2. Then the methodology to extract biomedical terms is detailed in Sect. 3. The results are presented in Sect. 4, followed by discussions in Sect. 5, and finally, the conclusions in Sect. 6.
2 Related work
Recent studies have focused on multi-word (n-grams) and single-word (unigrams) term extraction. Term extraction techniques can be divided into four broad categories: (i) Linguistic, (ii) Statistical, (iii) Machine Learning, and (iv) Hybrid. All of these techniques are encompassed in Text Mining approaches. Graph-based approaches have not yet been applied to ATE, although they have been successively adopted in other information retrieval fields and could be suitable for our purpose. Existing web techniques have not been applied to ATE but, as we will see, these techniques can be adapted for such purposes.
2.1 Text mining approaches
2.1.1 Linguistic approaches
These techniques attempt to recover terms via linguistic pattern formation. This involves building rules to describe naming structures for different classes based on orthographic, lexical, or morphosyntactic characteristics, e.g. Gaizauskas et al. 2000. The main approach is to develop rules (typically manually) describing common naming structures for certain term classes using orthographic or lexical clues, or more complex morpho-syntactic features. Moreover, in many cases, dictionaries of typical term constituents (e.g. terminological heads, affixes, and specific acronyms) are used to facilitate term recognition (Krauthammer and Nenadic December 2004). A recent study on biomedical term extraction (Golik et al. 2013) is based on linguistic patterns plus additional context-based rules to extract candidate terms, which are not scored and the authors leave the term relevance decision to experts.
2.1.2 Statistical methods
Statistical techniques chiefly rely on external evidence presented through surrounding (contextual) information. Such approaches are mainly focused on the recognition of general terms (Eck et al. 2010). The most basic measures are based on frequency. For instance, term frequency (tf) counts the frequency of a term in the corpus, document frequency (df) counts the number of documents where a term occurs, and average term frequency (atf), which is \(\frac{tf}{df}\).
A similar research topic, called automatic keyword extraction (AKE), proposes to extract the most relevant words or phrases in a document using automatic indexation. Keywords, which we define as a sequence of one or more words, provide a compact representation of a document’s content. Such measures can be adapted to extract terms from a corpus as well as ATE measures. We take two popular AKE measures as baselines measures, i.e. Term Frequency Inverse Document Frequency (TF-IDF) (Salton and Buckley 1988), and Okapi BM25 (Robertson et al. 1999) (hereafter Okapi), these weight the word frequency according to their distribution along the corpus. Residual inverse document frequency (RIDF) compares the document frequency to another chance model where terms with a particular term frequency are distributed randomly throughout the collection, while Chi-square (Matsuo and Ishizuka 2004) assesses how selectively words and phrases co-occur within the same sentences as a particular subset of frequent terms in the document text. This is applied to determine the bias of word co-occurrences in the document text, which is then used to rank words and phrases as keywords of the document; RAKE (Rose et al. 2010) hypothesises that keywords usually consist of multiple words and do not contain punctuation or stop words. It uses word co-occurrence information to determine the keywords.
2.1.3 Machine learning
Machine Learning (ML) systems are often designed for specific entity classes and thus integrate term extraction and term classification. Machine Learning systems use training data to learn features useful for term extraction and classification. But the availability of reliable training resources is one of the main problems. Some proposed ATE approaches use machine learning (Conrado et al. 2013; Zhang et al. 2010; Newman et al. 2012). However, ML may also generate noise and silence. The main challenge is how to select a set of discriminating features that can be used for accurate recognition (and classification) of term instances. Another challenge concerns the detection of term boundaries, which are the most difficult to learn.
2.1.4 Hybrid methods
Most approaches combine several methods (typically linguistic and statistically based) for the term extraction task. GlossEx (Kozakov et al. 2007) considers the probability of a word in the domain corpus divided by the probability of the appearance of the same word in a general corpus. Moreover, the importance of the word is increased according to its frequency in the domain corpus. Weirdness (Ahmad et al. 1999) considers that the distribution of words in a specific domain corpus differs from that in a general corpus. C/NC-value (Frantzi et al. 2000) combines statistical and linguistic information for the extraction of multi-word and nested terms. This is the most well-known measure in the literature. While most studies address specific types of entities, C/NC-value is a domain-independent method. It has also been used for recognizing terms in the biomedical literature (Hliaoutakis et al. 2009; Hamon et al. 2014). In (Zhang et al. 2008), the authors showed that C-value obtains the best results compared to the other measures cited above. C-value has been also modified to extract single-word terms (Nakagawa and Mori 2002), and in this work the authors extract only terms composed of nouns. Moreover, C-value has also been applied to different languages other than English, e.g. Japanese, Serbian, Slovenian, Polish, Chinese (Ji et al. 2007), Spanish (Barrón-Cedeño et al. 2009), Arabic, and French. We have thus chosen C-value as one of our baseline measure. Those baseline measures will be modified and evaluated with the new proposed measures.
Terminology extraction from parallel and comparable corpora Another kind of approach suggests that terminology may be extracted from parallel and/or comparable corpora. Parallel corpora contain texts and their translation into one or more languages, but such corpora are scarce (Bowker and Pearson 2002). Thus parallel corpora are scarce for specialized domains. Comparable corpora are those which select similar texts in more than one language or variety (Déjean and Gaussier 2002). Comparable corpora are built more easily than parallel corpora. They are often used for machine translation and their approaches are based on linguistics, statistics, machine learning, and hybrid methods. The main objective of these approaches is to extract translation pairs from parallel/comparable corpora. Different studies propose translation of biomedical terms for English-French by alignment techniques (Deléger et al. 2009). English–Greek and English–Romanian bilingual medical dictionaries are also constructed with a hybrid approach that combines semantic information and term alignments (Kontonatsios et al. 2014b). Other approaches are applied for single- and multi-word terms with English–French comparable corpora (Daille and Morin 2005). The authors use statistical methods to align elements by exploiting contextual information. Another study proposes to use graph-based label propagation (Tamura et al. 2012). This approach is based on a graph for each language (English and Japanese) and the application of a similarity calculus between two words in each graph. Moreover, some machine learning algorithms can be used, e.g. the logistic regression classifier (Kontonatsios et al. 2014a). There are also approaches that combine both corpora (Morin and Prochasson 2011) (i.e. parallel and comparable) in an approach to reinforce extraction. Note that our corpora are not parallel and are far of being comparable because of the difference in their size. Therefore these approaches are not evaluated in our study.
2.1.5 Tools and applications for biomedical term extraction
There are several applications implementing some measures previously mentioned, especially C-value for biomedical term extraction. The study of related tools revealed that most existing systems that especially implement statistical methods are made to extract keywords and, to a lesser extent, to extract terminology from a text corpus. Indeed, most systems take a single text document as input, not a set of documents (as corpus), for which the IDF can be computed. Most systems are available only in English and the most relevant for the biomedical domain are:
-
TerMine Footnote 2, developed by the authors of the C-value method, only for English term extraction;
-
Java Automatic Term Extraction Footnote 3 (Zhang et al. 2008), a toolkit which implements several extraction methods including C-value, GlossEx, TermEx and offer other measures such as frequency, average term frequency, IDF, TF-IDF, RIDF;
-
FlexiTerm Footnote 4 (Spasic et al. 2013), a tool explicitly evaluated on biomedical copora and which offer more flexibility than C-value when comparing term candidates (treating them as bag of words and ignoring the word order);
-
BioYaTeA Footnote 5 (Golik et al. 2013), is a version of the YaTeA term extractor (Aubin and Hamon 2006), both are available as a Perl module. It is a biomedical term extractor. The method used is based only on linguistic aspects.
-
BioTex Footnote 6 (Lossio-Ventura et al. 2014a), only for biomedical terminology extraction. It is available for online testing and assessment but can also be used in any program as a Java library (POS tagger not included). In contrast to other existing systems, this system allows us to analyze French and Spanish corpora, manually validate extracted terms and export the list of extracted terms.
2.2 Graph-based approaches
Graph modeling is an alternative for representing information, which clearly highlights relationships of nodes among vertices. It also groups related information in a specific way, and a centrality algorithm can be applied to enhance their efficiency. Centrality in a graph is the identification of the most important vertices within a graph. A host of measures have been proposed to analyze complex networks, especially in the social network domain (Borgatti 2005; Borgatti et al. 2009; Banerjee et al. 2014). Freeman (1979), formalized three different measures of node centrality: degree, closeness and betweenness. Degree is the number of neighbors that a node is connected to. Closeness is the inverse sum of shortest distances to all other neighbor nodes. Betweenness is the number of shortest paths from all vertices to all others that pass through that node. One study proposes to take the number of edges and their weights into account (Opsahl et al. 2010), since the three last measures do not do this. Another well known measure is PageRank (Page et al. 1999), which ranks websites. Boldi and Vigna (2014), evaluated the behavior of ten measures, and associated the centrality to the node with largest degree. Our approach proposes the opposite, i.e. we focus on nodes with a lower degree. An increasingly popular recent application of graph approaches to IR concerns social or collaborative networks and recommender systems (Noh et al. 2009; Banerjee et al. 2014).
Graph representations of text and scoring function definition are two widely explored research topics, but few studies have focused on graph-based IR in terms of both document representation and weighting models (Rousseau and Vazirgiannis 2015). First, text is modeled as a graph where nodes represent words and edges represent relations between words, defined on the basis of any meaningful statistical or linguistic relation (Blanco and Lioma 2012). In Blanco and Lioma (2012), the authors developed a graph-based word weighting model that represents each document as a graph. The importance of a word within a document is estimated by the number of related words and their importance, in the same way that PageRank (Page et al. 1999) estimates the importance of a page via the pages that are linked to it. Another study introduces a different representation of document that captures relationships between words by using an unweighted directed graph of words with a novel scoring function (Rousseau and Vazirgiannis 2015).
In the above approaches, graphs are used to measure the influence of words in documents like automatic keyword extraction methods (AKE), while ranking documents against queries. These approaches differ from ours as they use graphs focused on the extraction of relevant words in a document and computing relations between words. In our proposal, a graph is built such that the vertices are multi-word terms and the edges are relations between multi-word terms. Moreover, we focus especially on a scoring function of relevant multi-word terms in a domain rather than in a document.
2.3 Web mining approaches
Different web mining studies focus on semantic similarity, semantic relatedness. This means quantifying the degree to which some words are related, considering not only similarity but also any possible semantic relationship among them. The word association measures can be divided into three categories (Chaudhari et al. 2011): (i) co-occurrence measures that rely on co-occurrence frequencies of both words in a corpus, (ii) distributional similarity-based measures that characterize a word by the distribution of other words around it, and (iii) knowledge-based measures that use knowledge-sources like thesauri, semantic networks, or taxonomies (Harispe et al. 2014). In this paper, we focus on co-occurrence measures because our goal is to extract multi-word terms and we suggest computing a degree of association between words composing a term. Word association measures are used in several domains like ecology, psychology, medicine, and language processing, and were recently studied in (Pantel et al. 2009; Zadeh and Goel 2013), such as Dice, Jaccard, Overlap, Cosine. Another measure to compute the association between words using web search engines results is the Normalized Google Distance (Cilibrasi and Vitanyi 2007), which relies on the number of times words co-occur in the document indexed by an information retrieval system. In this study, experimental results with our web-based measure will be compared with the basic measures (Dice, Jaccard, Overlap, Cosine).
3 Methodology
This section describes the baseline measures, their modifications as well as new measures that we propose for the biomedical term extraction task. The principle of our approach is to assign a weight to a term, which represents the appropriateness of being a relevant biomedical term. This allows to give as output a list ranked by their appropriateness. Our methodology for automatic term extraction has three main steps plus an additional step (a), described in Fig. 1, and in the sections hereafter:
(a) Pattern construction,
(1) Candidate term extraction,
(2) Ranking of candidate terms,
(3) Re-ranking.
3.1 Pattern construction (step a)
As previously cited, we supposed that biomedical terms have a similar syntactic structure (linguistic aspect). Therefore, we built a list of the most common linguistic patterns according to the syntactic structure of terms present in the UMLSFootnote 7 (for English and Spanish), and the French version of MeSH,Footnote 8 SNOMED International and the rest of the French content in the UMLS.
Part-of-Speech (POS) tagging is the process of assigning each word in a text to its grammatical category (e.g. noun, adjective). This process is performed based on the definition of the word or on the context in which it appears. This is highly time-consuming, so we conducted automatic part-of-speech tagging.
We evaluated three tools (TreeTagger,Footnote 9 Stanford Tagger,Footnote 10 and Brill’s taggerFootnote 11). This evaluation was carried out throughout the entire workflow with the three tools and we assessed the precision of the extracted terms. We noted that in general TreeTagger gave the best results for Spanish and French. Meanwhile, for English, the Stanford tagger and TreeTagger gave similar results. We finally chose TreeTagger, which gave better results and may be used for English, French and Spanish. Moreover, our choice was validated with regard to a recent comparison study (Tian and Lo 2015), wherein the authors showed that TreeTagger generally gives the best results, particularly for nouns and verbs.
Therefore, we carried out automatic part-of-speech tagging of the biomedical terms using TreeTagger, and then computed the frequency of the syntactic structures. Patterns among the 200 highest frequencies were selected to build the list of patterns for each language. From this list, we also computed the weight (probability) associated with each pattern, i.e. the frequency of the pattern over the sum of frequencies (see Algorithm 1), but this weight will only be used for one measure. The number of terms used to build these lists of patterns was 3,000,000 for English, 300,000 for French, and 500,000 for Spanish, taken from the previously mentioned terminologies. Table 1 illustrates the computation of the linguistic patterns and their weights for English.
Different terminology extraction studies are based on the use of regular expressions to extract candidate terms, for instance (Frantzi et al. 2000). Generally these regular expressions are manually built for a specific language and/or domain (Daille et al. 1994). In our setting, we prefer to (i) construct and (ii) apply patterns in order to extract terms in texts. These patterns have the advantage of being generic because they are based on defined PoS tags. Moreover, they are very specific because they are (automatically) built with specialized biomedicine resources. Concerning this last point, we can consider we are close to the use of regular expressions. There are two main reasons that we use specific linguistic patterns. First, we would like to restrict the patterns to the biomedical domain. For instance, biomedical terms often contain numbers in their syntactic structure, and this is very specific to the biomedical domain, e.g. “epididymal protein 9”, “pargyline 10 mg”. General patterns do not enable extraction of such terms. Our methodology is based on 200 significant patterns for English, French, or Spanish, yet different for each language. For instance, there are 55 patterns for English that contain numbers in the linguistic structure. Thus, this kind of pattern seems quite relevant for this domain. The second reason for using lexical patterns is that we assign a probability of occurrence to each pattern, which would not be possible with classical patterns and regular expressions.

3.2 Candidate term extraction (step 1)
The first main step is to extract the candidate terms. So we apply part-of-speech to the whole corpus using TreeTagger. Then we filter out the content of our input corpus using previously computed patterns. We select only terms whose syntactic structure is in the patterns list. The pattern filtering is specifically done on a per-language basis (i.e. when the text is in French, only the French list of patterns is used).
3.3 Ranking of candidate terms (step 2)
We need to select the most appropriate terms for the biomedical domain. Candidate term ranking is therefore essential. For this purpose, several measures are proposed and Fig. 1(2) shows the set of available measures. We propose some modifications of the most known measures in the literature (i.e. C-value, TF-IDF, Okapi), and propose new ones (i.e. F-TFIDF-C, F-OCapi, LIDF-value, L-value). Those measures are linguistic- and statistic- based, they are also not very time-consuming. In this step, only one measure will be selected to perform the ranking. The measures of this section take a list of candidate terms previously filtered by linguistic patterns as input, which makes it possible to assess less invalid terms while dealing with the noise problem. In addition to the use of linguistic patterns to alleviate the problem of the extraction of multi-word terms having various complex structures. Moreover, the frequency decreases the number of invalid terms to evaluate (noise). The measures mentioned above are effective on large amounts of data (Lv and Zhai 2011a, b; Singhal et al. 1996), which overcomes the problem of large-scale corpora. Hereafter we describe all measures.
3.3.1 C-value
The C-value method combines linguistic and statistical information (Frantzi et al. 2000). Linguistic information is the use of a general regular expression as linguistic patterns, and the statistical information is the value assigned with the C-value measure based on the frequency of terms to compute the termhood (i.e. the association strength of a term to domain concepts). The C-value method aims to improve the extraction of long terms, and it was specially built for extracting multi-word terms.
where A is the candidate term, \(w(A) = \log _2(|A|)\), |A| the number of words in A, f(A) the frequency of A in the unique document, \(S_A\) the set of terms that contain A and \(|S_A|\) the number of terms in \(S_A\). In a nutshell, C-value uses either the frequency of the term if the term is not included in other terms (first line), or decreases this frequency if the term appears in other terms, based on the frequency of those other terms (second line).
We modified the measure in order to extract all terms (single-word + multi-words terms), as also suggested in (Barrón-Cedeño et al. 2009), but in a different manner.
The original C-value defines \(w(A) = \log _2(|A|)\), and we modified \(w(A) = \log _2(|A|+ {1} )\) in order to avoid null values for single-word terms, as illustrated in Table 2. Note that we do not use a stop word list or a frequency threshold as was originally proposed.
3.3.2 TF-IDF and Okapi
These measures are used to associate a weight to each term in a document (Salton and Buckley 1988). This weight represents the term relevance for the document. The output is a ranked list of terms for each document, which is often used in information retrieval so as to order documents by their importance for a given query (Robertson et al. 1999). Okapi can be seen as an improvement of the TF-IDF measure, while taking the document length into account.
where A is a term, considering d a document, D the collection of documents, f(A, d) the frequency of A in d, tf(A, d) the term frequency of A in d, idf(A, D) the inverse document frequency of A in D, dc(t) the number of documents containing term A, this means: \(|\{d \in D: t \in d \}|\), dl(d) the length of the document d in number of words, \(dl_{avg}\) the average document length of the collection.
As the output is a ranked list of terms per document, we could find the same term in different documents, with different weights in each document. So we need to merge the term into a single list. For this, we propose to merge them according to three functions, which respectively calculate the sum(S), max(M) and average(A) of the weights of a term. At the end of this task, we have three lists from Okapi and three lists from TF-IDF. The notation for these lists are \(Okapi_X(A)\) and \(\textit{TF}\hbox {-}\textit{IDF}_X(A)\), where A is the term, and X the factor \(\in \{M,S,A\}\). For example, \(Okapi_M(A)\) is the value obtained by taking the maximum Okapi value for a term A in the whole corpus. Figure 2 shows the merging process.
With aim of improving the term extraction precision, we designed two new combined measures, while taking the values obtained in the above steps into account. Both are based on harmonic means of two values.
3.3.3 Combinations: F-OCapi and F-TFIDF-C
Considered as the harmonic mean of the two used values, this method has the advantage of using all values of the distribution.
3.3.4 LIDF-value and L-value
In this section we present two new measures. The first one, called LIDF-value (Linguisitic patterns, IDF, and C-value information). LIDF-value is partially presented in Lossio-Ventura et al. (2014c). This is a new ranking measure based on linguistic and statistical information.
Our method LIDF-value is aimed at computing the termhood for each term, using the linguistic information calculated as described below, the idf, and the C-value of each term. The linguistic information gives greater importance to the term unithood in order to detect low frequency terms. So we associate the pattern weight (see Table 1) with the candidate term probability. That means the probability of a candidate term of being a relevant biomedical term. The probability is associated only if the syntactic structure of the term appears in the linguistic pattern list.
The inverse document frequency (idf) is a measure indicating the extent to which a term is common or rare across all documents. It is obtained by dividing the total number of documents by the number of documents containing the term, and then by taking the logarithm of that quotient. The probability and idf improve low frequency term extraction. The objective of these two components is to tackle the silence problem, allowing extraction of discriminant terms, for instance, in a biomedical corpus, “virus production” with low frequency being better ranked than “human monocytic cell”, which has a higher frequency. This means that for a low frequency candidate term, its score can be favored if its linguistic pattern is associated with a high probability and/or its idf value is also high. The C-value measure is based on the term frequency. The C-value (see formula 1) measure favors a candidate term that does not often appear in a longer term. For instance, in a specialized corpus (ophthalmology), the authors of Frantzi et al. (2000) found the irrelevant term “soft contact” while the frequent and longer term “soft contact lens” is relevant.
As an example, we implement the Algorithm 2, which describes the applied process. These different statistical information items (i.e. probability of linguistic patterns, C-value, idf) are combined to define the global ranking measure LIDF-value (see formula 6); where \({\rm {P}}(A_{LP})\) is the probability of a term A which has the same linguistic structure pattern LP, i.e. the weight of the linguistic pattern LP computed in Subsection Pattern Construction.

Note that LIDF-value works only for a set of documents, mainly because the idf measure can only be computed on a set of documents (see formula 2). Therefore, for datasets composed of one document, we propose a new measure, L-value, as explained in the following paragraphs.
L-value is a variant of LIDF-value, focused on one document with the goal of benefiting from the probability of linguistic patterns computed for LIDF-value. This measure does not contain the idf (see formula 7). L-value is interesting to highlight the more representative terms of a single corpus without considering the discriminative aspects, e.g. idf. This measure gives another point of view and is complementary to those based on the idf weighting.
A single document can be considered as a free text without delimitation. For instance, a scientist article, a book, a document created with titles/abstracts from a library database. L-value becomes interesting when it does not exist a considerable amount of data for a new subject, i.e. an emergent term in the community. For instance, the “Ataxia Neuropathy Spectrum” term appears only in four titles/abstracts of scientist articles from PubMedFootnote 12 between 2009 and 2015. PubMed is a free search engine accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics.
3.4 Re-ranking (step 3)
After the term extraction, we propose new measures to re-rank the candidate terms in order to increase the top k term precision. The re-ranking measures aim to improve the term extraction results of ranking measures. This involves positioning the most relevant biomedical terms at the top of the list. That provides more confidence that the terms appearing at the top of this list are true biomedical terms.
These re-ranking functions represent an extension of the measures presented in Lossio-Ventura et al. (2014b). Therefore, as improvements, we propose to take graph-theoretic information into account to highlight relevant terms, as well as web information, as explained in the following subsections. These measures can be executed separately, but the graph construction is time consuming, and the number of search engine queries is limited. Therefore, we just apply these measures for a group of selected terms given by a ranking measure. Because the ranking measures have proved to be more efficient applied before than TeRGraph and web-based measures.
As these measures are applied to the list of terms obtained with a ranking measure, which tackles noise, silence and multi-word term extraction problems, so they also take into account those problems. As mentioned, the objective of re-raking measures is to re-rank terms, so the manual validation efforts of the candidate terms decrease because the relevant biomedical term is allocated at the top of the list.
3.4.1 A new graph-based ranking measure: “TeRGraph” (terminology ranking based on graph information)
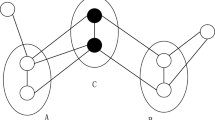
This approach aims to improve the ranking (and therefore the precision results) of extracted terms. As mentioned above, in contrast to the above-cited study, the graph is built with a list of terms obtained according to a measure described in Sect. 3.2, where vertices denote terms linked by their co-occurrence in sentences in the corpus. Moreover, we make the hypothesis that the term representativeness in a graph, for a specific-domain, depends on its number of neighbors, and the number of neighbors of its neighbors. We assume that a term with more neighbors is less representative of the specific domain. This means that this term is used in the general domain. Figure 3 illustrates our hypothesis.
The graph-based approach is divided into two steps:
-
(i)
Graph construction a graph (see Fig. 5) is built where vertices denote terms, and edges denote co-occurrence relations between terms, co-occurrences between terms are measured as the weight of the relation in the initial corpus. This approach is statistical because it links all co-occurring terms without considering their meaning or function in the text. This graph is undirected as the edges imply that terms simply co-occur, without any further distinction regarding their role. We take the Dice coefficient, a basic measure to compute the co-occurrence between two terms x and y, as defined by the following formula:
$$D(x,y)= {} \frac{2 \times P(x,y)}{P(x)+P(y)}$$(8) -
(ii)
Representativeness computations on the term graph a principled graph-based measure to compute term weights (representativeness) is defined. The aim of this new graph-based ranking measure, TeRGraph, see Eq. 9, is to derive these weights for each vertex, (i.e. multi-word term weight), in order to re-rank the list of extracted terms.
where A represents a vertex (term), \({\rm {N}}(A)\) the neighborhood of A, \(|{\rm {N}}(A)|\) the number of neighbors of A, \(T_i\) the neighbor i of A, and k a constant. The intuition for Eq. 9 is as follows: the more a term A has neighbors (directly with \({\rm {N}}(A)\) or by transitivity with \({\rm {N}}(T_i)\)), the more the weight decreases. Indeed, a term A having a lot of neighbors is considered too general for the domain (i.e. this term is not salient), so it has to be penalized via the associated score.
The k constant affects the TeRGraph value, i.e. the set of values that TeRGraph takes when k changes. For instance, when \(k = 0.5\), the set of values for TeRGraph is between −1 and 0, (i.e. \(TeRGraph \in [-1,0]\)), and when \(k = 1\), \(TeRGraph \in [0,0.6]\). As the values taken by TeRGraph are different, then the slope of the curve is also different. Figure 4 shows the behavior of TeRGraph when k changes. According the experiments, we have chosen \(k=1.5\). The main reason is that the slope of the curve is low, and the set of values for TeRGraph ranges from 0.6 to 1.
See Algorithm 3 for more details, it describes the entire process: (1) co-occurrence graph construction, (2) computation of the representativeness of each vertex.

Figure 5 shows an example to calculate the value of TeRGraph for a term in different graphs. These graphs are built with different co-occurrence thresholds (i.e. Dice’s value between two terms). In this example, \(A_1\) and \(A_2\) represent the term chloramphenicol acetyltransferase reporter in Graphs 1 and 2, respectively.
3.4.2 WebR
The aim of our web-based measure, to predict with a better confidence if a candidate term is a valid biomedical term or not. It is appropriated for multi-word terms, as it computes the dependence between the words of a term. In our case, we compute a “strict” dependence, which means the proximity of words of terms (i.e. neighboring words) is calculated with a strict restriction. In comparison to other web-based measures (Cilibrasi and Vitanyi 2007), WebR reduces the number of pages to consider by taking only web pages containing all words of the terms into account. In addition, our measure can be easily adopted for all types of multi-word terms.
where \(A =\) multi-word term, \(a_i \in A\) and \(a_i = \{noun,\) adjective, foreign \(word\}\). Where A is the candidate term, \(nb(\hbox {``}A\hbox {''})\) the number of hits returned by a web search engine with exact match only with multi-word term A (query with quotation marks “A”), nb(A) the number of documents returned by the search engine, including not exact matches (query A without quotation marks), i.e. whole documents containing words of the multi-word term A. For example, the multi-word term treponema pallidum, will generate two queries, the first \(nb(\hbox {``} treponema \, pallidum \hbox {''})\) which returns with Yahoo 1,100,000 documents, and the second query \(nb( treponema \, pallidum )\) which returns 1,300,000 documents, then \(\textit{WebR(treponema pallidum)}= \frac{1100000}{1300000} = 0.85\).
In our workflow, we tested Yahoo and Bing. WebR re-rank the list of candidate terms returned by the combined measures.
3.4.3 A new web ranking measure: WAHI (Web Association based on Hits Information)
Previous studies of web mining approaches query the web via search engines to measure word associations. This enables measurement of the association of words composing a term (e.g. soft, contact, and lens that compose the relevant term soft contact lens). To measure this association, our web-mining approach takes the number of pages provided by search engines into account (i.e. number of hits).
Our web-based measure re-ranks the list obtained previously with TeRGraph. We will show that this improves the precision of the k first terms extracted (see Sect. 4) and that it is specially appropriate for multi-word term extraction.
Formula 8 leads directly to formula 11.Footnote 13 The nb function used in formula 11 represents the number of pages returned by search engines (i.e. Yahoo and Bing). With this measure, we compute a strict dependence (i.e. neighboring words by using the operator ’ ” ’ of search engines). For instance, x might represent the word soft and y the word contact in order to calculate the association measure of the soft contact term.
Then we extend this formula to n elements as follows:
This measure enables us to calculate a score for all multi-word terms, such as soft contact lens.
To obtain WAHI, we propose to associate Dice criteria with WebR (see formula 10). This only takes the number of web pages containing all the words of the terms into account by using operators “ ” and AND.
For example, soft contact lens, the numerator corresponds to the number of web pages with the query “soft contact lens”, and for the denominator, we consider the query soft AND contact AND lens.
Finally, the global ranking approach combining Dice and WebR is given by WAHI measure (Web Association based on Hits Information):
Algorithm 4 details the global web mining process to rank terms. We show in the next section that open-domain (general) resources, such as the web, can be tapped to support domain-specific term extraction. They can thus be used to compensate for the unavailability of domain-specific resources.

4 Experiments and results
4.1 Data, protocol, and validation
4.1.1 Data
We used two corpora for our experiments. The first one is a set of biological laboratory tests, extracted from LabTestsOnline.Footnote 14 This website provides information in several languages to patients or family caregivers about clinical lab tests. Each test that forms a document in our corpus includes the formal lab test name, some synonyms and possible alternate names as well as a description of the test. The LabTestsOnline website was extracted totally for English, French, and Spanish with a crawler created specifically for this purpose. These documents are available online.Footnote 15 Table 3 shows the details of LabTestsOnline corpus for different languages.
The second corpus is GENIA,Footnote 16 which is made up of 2000 titles and abstracts of journal articles that were culled from the Medline database, with more than 400,000 words in English. The GENIA corpus contains linguistic expressions referring to entities of interest in molecular biology, such as proteins, genes and cells. GENIA is an annotated dataset, in which technical term annotation covers the identification of physical biological entities as well as other important terms. This is our gold standard corpus. Whereas the Medline indexes a broad range of academic articles covering the general or specific domains of life sciences, GENIA is intended to cover a smaller subject domain: biological reactions concerning transcription factors in human blood cells.
4.1.2 Protocol
As the measures described in step 2 of our workflow (i.e. Ranking the Candidate Terms) are not very time-consuming, and as they are easily applicable for large corpora, they were evaluated over the LabTestsOnline corpus for English, French, and Spanish, and over the gold standard corpus, GENIA. In contrast, as the measures described in step 3 (i.e. Re-ranking) are highly time-consuming, and they are used at the end of the process, to enhance the performance of the results, we evaluate them only over the GENIA corpus.
4.1.3 Validation
In order to automatically validate and cover medical terms, we use UMLS for English and Spanish, and the French version of MeSH, SNOMED International and the rest of the French content in the UMLS. For instance, if an extracted candidate term is found in the UMLS dictionary, this term will be automatically validated. The results are evaluated in terms of precision obtained over the top k extracted terms (P@k).
Biomedical terminologies or ontologies (e.g. UMLS, SNOMED, MeSH), contain terms composed of signs. Therefore, we cleaned these terminologies by eliminating all terms containing (; , ? ! : { } [ ]), and we only took terms without signs. Table 4 shows the distribution in n-gram (i.e. n-gram is a term of n words, with \(n \ge 1\)) of biomedical resources for three languages, as well as the number of terms that we took after the cleaning task. For instance, the first cell means that 13.73 % of terms are composed of one word (1-gram) in UMLS for English.
4.2 Multilingual comparison (LabTestsOnline)
In this section, we show results obtained only with all the ranking measures, i.e. step 2 (ranking) in Fig. 1. In addition, we tested the measures for single- plus multi-word terms, or just for multi-word terms in English, French and Spanish. Tables 5, 6 and 7 show the results in English, French and Spanish, respectively. At the top of each table, the \(\hbox {single-word} + \hbox {multi-word}\) term extraction results are presented, while the multi-word term extraction results are presented at the bottom of the table.
These tables show that LIDF-value and L-value obtain the best results for both extraction cases and for the three languages. The combined measures based on the harmonic mean, and on the SUM and MAX (i.e. \(F \hbox {-} TFIDF \hbox {-} C _M\), \(F \hbox {-} TFIDF \hbox {-} C _S\)), also give interesting results.
The \(\hbox {single-word} + \hbox {multi-word}\) term extraction results are better than just the multi-word term extraction results. The main reason for this is that the extraction of single-word terms is more efficient due to their syntactic structure (linguistic structure), i.e. usually a noun. In addition, this syntactic structure has fewer variations. The results are lower as compared to multi-word term extraction, which is more complicated and involves more variations.
We observe that LIDF-value and L-value obtain very close results. In most cases LIDF-value performs better than L-value. These two measures show that the probability associated with the linguistic patterns helps to improve the term extraction results. Note that the idf influences LIDF-value, for this reason LIDF-value has better results than L-value.
4.3 Evaluation of the global process (GENIA)
Since GENIA is the gold standard corpus, we conduct a detailed assessment of the experiments in this subsection. We evaluated the entire workflow of our methodology, i.e. steps 2 (ranking) and 3 (re-ranking) in Fig. 1. As noted earlier, the multi-word term extraction results are influenced by the syntactic structure and their variations. So our experimentation in this subsection is focused only on multi-word term extraction.
In the following paragraphs, we also narrow down the presented results by keeping only the first 8000 extracted terms for the graph-based measure and the first 1000 extracted terms for the web-based measure.
4.3.1 Ranking results (step 2 in Fig. 1)
Table 8 presents and compares the multi-word term extraction results with the best ranking measures, as shown earlier, i.e. C-value, \(F \hbox {-} TFIDF \hbox {-} C _M\), and LIDF-value. The best results were obtained with LIDF-value with an 11 % improvement in precision for the first hundred extracted multi-word terms. These precision results are also shown in Fig. 6. The precision of LIDF-value will be further improved with TeRGraph.
4.3.2 Results of n-gram terms
We also evaluated C-value, \(F \hbox {-} TFIDF \hbox {-} C _M\), and LIDF-value in a sequence of n-gram terms (i.e. n-gram term is a multi-word term of n words), for this we require an index term to be a n-gram terms of length \(n \ge 2\). We tested the performance of LIDF-value on the n-gram term extraction taking the first 1000 n-g terms (\(n \ge 2\)).
Table 9 shows the precision comparison for the 2-gram, 3-gram and \(4+\) gram term extracted with C-value, \(F \hbox {-} TFIDF \hbox {-} C _M\), and LIDF-value. We can see that LIDF-value obtains the best results for all intervals for any \(n \ge 2\). These precision results are also shown in Fig. 7 for the 2-gram terms, Fig. 8 for the 3-gram terms, and finally Fig. 9 for the \(4+\) gram terms.
Table 10 shows the top-20 ranked 2-gram terms extracted with the baseline measures and LIDF-value. C-value obtained three irrelevant terms, F-TFIDF-C obtained five irrelevant terms while LIDF-value obtained only two irrelevant terms for the top-20 ranked 2-gram terms.
Similarly, Table 11 shows top-10 ranked 3-gram terms extracted with the baseline measures and LIDF-value. Finally, Table 12 shows the top-10 ranked \(4+\) gram terms extracted with the baseline measures and LIDF-value.
Note that in this context, “irrelevant” means that the terms are not in the above mentioned resources. These candidate terms might be interesting for ontology extension or population, however they must pass through polysemy detection in order to identify the possible meanings.
4.3.3 Re-ranking results (step 3 in Fig. 1)
Graph-based results our graph-based approach is applied to the first 8000 terms extracted by the best ranking measure. The objective is to re-rank the 8000 terms while trying to improve the precision by intervals. One parameter is involved in the computation of graph-based term weights, i.e. the threshold of Dice value which represents the relation when building the term graph. This involves linking terms whose Dice value of the relation is higher than threshold. We vary threshold (\(\delta\)) within \(\delta = [0.25, 0.35, 0.50, 0.60, 0.70]\) and report the precision performance for each of these values. Table 13 gives the precision performance obtained by TeRGraph and shows that it is well adapted for ATE.
Web-based results Our web-based approach is applied at the end of the process, with only the first 1000 terms extracted during the previous linguistic, statistic and graph measures. For space reasons, we show only the results obtained with WAHI, which are higher than WebR.
We took the list obtained with TeRGraph and \(\delta \ge 0.60\). The main reason for this limitation is the limited number of automatic queries possible in search engines. At this step, the aim is to re-rank the 1000 terms to try to improve the precision by intervals. Each measure listed in Table 14 and Table 15 shows the precision obtained after re-ranking. We tested WAHI with Yahoo and Bing search engines.
Table 14 and Table 15 prove that WAHI (either using Yahoo or Bing) is well adapted for ATE and this measure obtains better precision results than the baselines measures for word association. So our measures obtain real terms of our dictionary with a better ranking.
4.3.4 Summary
LIDF-value obtains the best precision results for multi-word term extraction, for each index term extraction (n-gram) and for intervals.
Table 16 presents a precision comparison of LIDF-value and TeRGraph measures. In terms of overall precision, our experiments produce consistent results from the GENIA corpus. In most cases, TeRGraph obtains better precision with a \(\delta\) of 0.60 and 0.70 (i.e. better precision in most P@k intervals), which is very good because it helps alleviate the problem of manual validation of candidate terms. These precisions are also illustrated in Fig. 10.
The performance of our graph-based measure somewhat depends on the value of the co-occurrence relation between terms. Specifically, the value of the co-occurrence relation affects how the graph is built (whose edges are taken), and hence it is critical for computation of the graph-based term weight. Another performance factor of our graph-based measure is the quality of the results obtained with LIDF-value due to the fact that the list of terms extracted with LIDF-value is required as input to re-rank TeRGraph in order to construct the graph, where nodes denote terms, and edges denote co-occurrence relations.
Table 17 presents the precision comparison of our three measures.
WAHI based on Yahoo obtains better precision for the first P@100 extracted terms with 96 % precision whereas, in comparison, WAHI based on Bing obtains 90 precision. For the other interval, Table 17 shows that WAHI based on Bing generally gives the best results. This is very encouraging because it also helps alleviate the problem of manual validation of candidate terms.
The performance of WAHI depends on the search engine because algorithms designed for searching information on the web are different, so the number of hits returned will differ in all cases. Another performance factor is the quality of the re-ranked list obtained with TeRGraph, because this list is required as input.
Moreover, Table 17 highlights that re-ranking with WAHI enables us to increase the precision of TeRGraph. For all cases, our re-ranking methods improve the precision obtained with LIDF-value. The purpose for which this web-mining measure was designed has thus been fulfilled.
Note that these measures do not normalize the possible variants. This could be a limitation for researchers looking for a preferred term for a group of variants.
5 Discussion
We discuss the effects of some parameters of our workflow. In the next sections, we explain the impacts of biomedical pattern lists, size of dictionaries, and the extraction errors.
5.1 Impact of pattern list
In our methodology, we have shown that biomedical patterns directly affect the term extraction results. For instance, we can see that L-value, which is a combination of C-value and the probability of pattern lists, gives better results than C-value for the three languages, and LIDF-value outperforms L-value in major cases. These pattern lists work specifically for the biomedical domain. If we use these biomedical patterns in another domain instead of using specific patterns of that domain, they will impact the term extraction results. To prove this, we have extracted terms from an agronomic corpus for English and French while taking biomedical patterns and agronomic patterns into account. We built the agronomic patterns using AGROVOC,Footnote 17 which is an agronomic dictionary. AGROVOC contains 39,542 and 37,382 English and French terms, respectively. Our corpus consists of titles plus abstracts extracted from the list of Cirad publications (French Agricultural Research Centre for International Development). Table 18 shows the details of the corpus formed for this comparison.
Table 19 presents a term extraction comparison while taking patterns built from two different domains into account. Again we note that LIDF-value obtains the best results. We also see that the results of terms extracted with agronomic patterns give better results than when using biomedical patterns for English and French.
Note that even if the term extraction results obtained using agronomic patterns are higher than using biomedical patterns, these results are a bit close. The main reason is that the biomedical and agronomic terms overlap. It means that identical patterns exist in both domains. The results could be improved by using patterns of two completely different domains.
5.2 Effect of dictionary size
Dictionaries play an important role in term extraction, specifically during the construction of pattern lists. Table 19 shows that a reduction in dictionary size degrades the performance of the precision results in comparison to Tables 5, 6, and 8. For instance, for the agronomic and biomedical domain, Tables 19 and 5 show the P@100 of 0.92 and 1.00 respectively, and this difference increases as the number of extracted terms increases (i.e. P@k).
5.3 Term extraction errors
As explained in Sect. 3 (step a), the term extraction results are influenced by the Part-of-Speech (PoS) tagging tools, which have different results for different languages. Briefly, the tool “A” can perform very well for English, while for French the tool “B” gives the best results. For instance, the sentence “Red blood cells increase with ...” was tagged with the Stanford tool as “adjective noun noun verb preposition ...”, whereas the TreeTagger tool tagged it as “adjective noun noun noun preposition ...”. Therefore, in order to show the generality of our approach, we choose a uniform PoS tool, i.e. TreeTagger, as a trade-off for three languages (English, French, and Spanish), while understanding that it will penalize the results for the three languages.
6 Conclusions and future work
This paper defines and evaluates several measures for automatic multi-word term extraction. These measures are classified as ranking measures, and re-ranking measures. The measures are based on the linguistic, statistical, graphic and web information. We modified some baseline measures (i.e. C-value, TF-IDF, Okapi) and we proposed new measures.
All the ranking measures are linguistic- and statistic-based. The best ranking measure is LIDF-value, which overcomes the lack of frequency information with the linguistic pattern probability and idf values.
We experimentally showed that LIDF-value applied in the biomedical domain, over two corpora (i.e. LabTestsOnline, GENIA), outperformed a state-of-the-art baseline for extracting terms (i.e. C-value), while obtaining the best precision results in all intervals (i.e. \(P\hbox {@}k\)). And with three languages the LIDF-value trends were similar.
We have shown that multi-word term extraction is more complex than single-word term extraction. We detailed an evaluation over the GENIA corpus for multi-word term extraction. Moreover, in that case, LIDF-value improved the automatic term extraction precision in comparison to the most popular term extraction measure.
We also evaluated the re-ranking measures. The first re-ranking measure, TeRGraph, is a graph-based measure. It decreases the human effort required to validate candidate terms. The graph-based measure has never been applied for automatic term extraction. TeRGraph takes the neighborhood to compute the term representativeness in a specific domain into account.
The other re-ranking measures are web-based. The best one, called WAHI, takes the list of terms obtained with TeRGraph as input. WAHI enables us to further reduce the huge human effort required for validating candidate terms.
Our experimental evaluations revealed that TeRGraph had better precision than LIDF-value for all intervals. Moreover, our experimental assessments revealed that WAHI improved the results given with TeRGraph for all intervals.
As a future extension of this work, we intend to use the relation value within TeRGraph. We plan to include the use of other graph ranking computations, e.g. PageRank, adapted for automatic term extraction. Moreover, a future work consists of using the web to extract more terms than those extracted.
One prospect could be the creation of a regular expression for the biomedical domain from the linguistic pattern list. We plan to modify our measures in order to normalize the possible variants, looking towards for a preferred term for those variants.
Notes
by writing \(P(x)=\frac{nb(x)}{nb\_total}\), \(P(y)=\frac{nb(y)}{nb\_total}\), \(P(x,y)=\frac{nb(x,y)}{nb\_total}\).
References
Ahmad, K., Gillam, L., & Tostevin, L. (1999). University of surrey participation in TREC-8: Weirdness indexing for logical document extrapolation and retrieval (wilder). In TREC.
Aubin, S., & Hamon, T. (2006). Improving term extraction with terminological resources. In Proceedings of the 5th international conference natural language processing (pp. 380–387). FinTAL’06 Turku, Finland: Springer.
Banerjee, A., Chandrasekhar, A. G., Duo, E., & Jackson, M. O. (2014). Gossip: Identifying central individuals in a social network. Technical report, National Bureau of EconomicResearch.
Barrón-Cedeño, A., Sierra, G., Drouin, P., & Ananiadou, S. (2009). An improved automatic term recognition method for spanish. In Proceedings of the 10th international conference on computational linguistics and intelligent text processing (pp. 125–136) CICLing’09. Springer.
Blanco, R., & Lioma, C. (2012). Graph-based term weighting for information retrieval. Information Retrieval, 15(1), 54–92.
Boldi, P., & Vigna, S. (2014). Axioms for centrality. Internet Mathematics, 10(3–4), 222–262.
Borgatti, S. P. (2005). Centrality and network flow. Social Networks, 27(1), 55–71.
Borgatti, S. P., Mehra, A., Brass, D. J., & Labianca, G. (2009). Network analysis in the social sciences. Science, 323(5916), 892–895.
Bowker, L., & Pearson, J. (2002). Working with specialized language: A practical guide to using corpora. London: Routledge.
Chaudhari, D. L., Damani, O. P., & Laxman, S. (2011). Lexical co-occurrence, statistical significance, and word association. In Proceedings of the conference on empirical methods in natural language processing (pp. 1058–1068). EMNLP’11, Stroudsburg, PA, USA. Association for Computational Linguistics.
Cilibrasi, R. L., & Vitanyi, P. M. (2007). The google similarity distance. IEEE Transactions on Knowledge and Data Engineering, 19(3), 370–383.
Conrado, M. S., Pardo, T. A., & Rezende, S. O. (2013). Exploration of a rich feature set for automatic term extraction. Advances in Artificial Intelligence and Its Applications (pp. 342–354), vol. 8265 of Lecture Notes in Computer Science Berlin Heidelberg: Springer.
Daille, B., Gaussier, E., & Langé, J.-M. (1994). Towards automatic extraction of monolingual and bilingual terminology. In Proceedings of the 15th conference on computational linguistics—Volume 1, COLING’94, pages 515–521, Stroudsburg, PA, USA. Association for Computational Linguistics.
Daille, B., & Morin, E. (2005). French-english terminology extraction from comparable corpora. In Proceedings of the 2nd international joint conference natural language processing (pp. 707–718). IJCNLP’05. Springer.
Déjean, H., & Gaussier, E. (2002). Une nouvelle approche à l’extraction de lexiques bilingues à partir de corpus comparables.
Deléger, L., Merkel, M., & Zweigenbaum, P. (2009). Translating medical terminologies through word alignment in parallel text corpora. Journal of Biomedical Informatics, 42(4), 692–701.
Dobrov, B., & Loukachevitch, N. (2011). Multiple evidence for term extraction in broad domains. In Proceeding of recent advances in natural language processing (pp. 710–715). RANLP’11 Bulgaria: Hissar.
Frantzi, K., Ananiadou, S., & Mima, H. (2000). Automatic recognition of multi-word terms: The c-value/nc-value method. International Journal on Digital Libraries, 3(2), 115–130.
Freeman, L. C. (1979). Centrality in social networks conceptual clarification. Social Networks, 1(3), 215–239.
Gaizauskas, R., Demetriou, G., & Humphreys, K. (2000). Term recognition and classification in biological science journal articles. In Proceeding of the computional terminology for medical and biological applications workshop of the 2nd international conference on NLP (pp. 37–44).
Golik, W., Bossy, R., Ratkovic, Z., & Nédellec, C. (2013). Improving term extraction with linguistic analysis in the biomedical domain. In Proceedings of the 14th international conference on intelligent text processing and computational linguistics, special issue of the journal Research in Computing Science (pp. 24–30). CICLing’13.
Hamon, T., Engström, C., & Silvestrov, S. (2014). Term ranking adaptation to the domain: Genetic algorithm-based optimisation of the c-value. In Proceedings of the 9th international conference on natural language processing (pp. 71–83). PolTAL’2014 - LNAI Warsaw, Poland: Springer.
Harispe, S., Ranwez, S., Janaqi, S., & Montmain, J. (2014). The semantic measures library and toolkit: Fast computation of semantic similarity and relatedness using biomedical ontologies. Bioinformatics, 30(5), 740–742.
Hliaoutakis, A., Zervanou, K., & Petrakis, E. G. (2009). The amtex approach in the medical document indexing and retrieval application. Data and Knowledge Engineering, 68(3), 380–392.
Ji, L., Sum, M., Lu, Q., Li, W., & Chen. Y. (2007). Chinese terminology extraction using window-based contextual information. In Proceedings of the 8th international conference on computational linguistics and intelligent text processing (pp. 62–74). CICLing’07, Berlin, Heidelberg. Springer-Verlag.
Kageura, K., & Umino, B. (1996). Methods of automatic term recognition: A review. Terminology, 3(2), 259–289.
Kontonatsios, G., Korkontzelos, I., Tsujii, J., & Ananiadou, S. (2014). Combining string and context similarity for bilingual term alignment from comparable corpora. In Proceedings of the 2014 conference on empirical methods in natural language processing (pp. 1701–1712). EMNLP’14, Doha, Qatar. Association for Computational Linguistics.
Kontonatsios, G., Mihăilă, C., Korkontzelos, I., Thompson, P., & Ananiadou, S. (2014). A hybrid approach to compiling bilingual dictionaries of medical terms from parallel corpora. In Statistical language and speech processing pp. 57–69. Springer.
Kozakov, L., Park, Y., Fin, T., Drissi, Y., Doganata, N., & Confino, T. (2007). Glossary extraction and knowledge in large organisations via semantic web technologies. In Proceedings of the 6th international semantic web conference and he 2nd Asian semantic web conference (semantic web challenge track), ISWC-ASWC’07. Springer.
Krauthammer, M., & Nenadic, G. (2004). Term identification in the biomedical literature. Journal of Biomedical Informatics, 37(6), 512–526.
Lossio-Ventura, J. A., Hacid, H., Ansiaux, A., & Maag, M. L. (2012). Conversations reconstruction in the social web. In Proceedings of the 21st international conference companion on World Wide Web (pp. 573–574). WWW’12, Lyon, France, ACM.
Lossio-Ventura, J. A., Jonquet, C., Roche, M., & Teisseire, M. (2014). BIOTEX: A system for biomedical terminology extraction, ranking, and validation. In Proceedings of the 13th international semantic web conference, posters and demonstrations track (pp. 157–160). ISWC’14.
Lossio-Ventura, J. A., Jonquet, C., Roche, M., Teisseire, M., & ACM. (2014). Integration of linguistic and web information to improve biomedical terminology extraction. In Proceedings of the 18th international database engineering and applications symposium (pp. 265–269). IDEAS’14 Porto, Portugal: ACM.
Lossio-Ventura, J. A., Jonquet, C., Roche, M., & Teisseire, M. (2014). Yet another ranking function for automatic multiword term extraction. In Proceedings of the 9th international conference on natural language processing, number 8686 in PolTAL’2014 - LNAI (pp. 52–64). Warsaw, Poland, Springer.
Lv, Y., & Zhai, C. (2011). Adaptive term frequency normalization for BM25. In Proceedings of the 20th ACM international conference on information and knowledge management (pp. 1985–1988). CIKM’11, New York, NY, USA. ACM.
Lv, Y., & Zhai, C. (2011). When documents are very long, BM25 fails! In Proceedings of the 34th international acm sigir conference on research and development in information retrieval, SIGIR’11 (pp. 1103–1104). New York, NY, USA. ACM.
Matsuo, Y., & Ishizuka, M. (2004). Keyword extraction from a single document using word co-occurrence statistical information. International Journal on Artificial Intelligence Tools, 13(01), 157–169.
Morin, E., & Prochasson, E. (2011). Bilingual lexicon extraction from comparable corpora enhanced with parallel corpora. In Proceedings of the 4th workshop on building and using comparable corpora: comparable corpora and the web (pp. 27–34). Association for Computational Linguistics.
Murdoch, T. B., & Detsky, A. S. (2013). The inevitable application of big data to health care. Journal of the American Medical Association, JAMA, 309(13), 1351–1352.
Nakagawa, H., & Mori, T. (2002). A simple but powerful automatic term extraction method. In COLING-02 on COMPUTERM 2002: Second International Workshop on Computational Terminology—Vol. 14, COMPUTERM ’02 (pp. 1–7). Stroudsburg, PA, USA, Association for Computational Linguistics.
Névéol, A., Grosjean, J., Darmoni, S. J., & Zweigenbaum, P. (2014). Language resources for french in the biomedical domain. In Proceedings of the 9th international conference on language resources and evaluation, LREC’14. Association for Computational Linguistics.
Newman, D., Koilada, N., Lau, J. H., & Baldwin, T. (December 2012). Bayesian text segmentation for index term identification and keyphrase extraction. In Proceedings of 24th international conference on computational linguistics (pp. 2077–2092). COLING’12 India: Mumbai.
Noh, T.-G., Park, S.-B., Yoon, H.-G., Lee, S.-J., & Park, S.-Y. (2009). An automatic translation of tags for multimedia contents using folksonomy networks. In Proceedings of the 32Nd international ACM SIGIR conference on research and development in information retrieval, SIGIR’09 (pp. 492–499). New York, NY, USA, ACM.
Noy, N. F., Shah, N. H., Whetzel, P. L., Dai, B., Dorf, M., Griffith, N. B., et al. (2009). Bioportal: Ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research, 37, 170–173.
Opsahl, T., Agneessens, F., & Skvoretz, J. (2010). Node centrality in weighted networks: Generalizing degree and shortest paths. Social Networks, 32(3), 245–251.
Page, L., Brin, S., Motwani, R., & Winograd, T. (1999). The pagerank citation ranking: Bringing order to the web.
Pantel, P., Crestan, E., Borkovsky, A., Popescu, A.-M., & Vyas, V. (2009). Web-scale distributional similarity and entity set expansion. In Proceedings of the conference on empirical methods in natural language processing, EMNLP’09 (pp. 938–947). Stroudsburg, PA, USA. Association for Computational Linguistics.
Qureshi, M. A., O’Riordan, C., & Pasi, G. (2012). Short-text domain specific key terms/phrases extraction using an n-gram model with wikipedia. In Proceedings of the 21st ACM international conference on information and knowledge management, CIKM’12 (pp. 2515–2518). New York, NY, USA, ACM.
Robertson, S. E., Walker, S., & Beaulieu, M. (1999). Okapi at TREC-7: Automatic ad hoc, filtering, vlc and interactive track. IN, 21, 253–264.
Rose, S., Engel, D., Cramer, N., & Cowley, W. (2010). Automatic keyword extraction from individual documents. In M. W. Berry, J. Kogan (Eds.), Text Mining: Applications and Theory (pp. 1–20). John Wiley and Sons, Ltd.
Rousseau, F., & Vazirgiannis, M. (2013). Graph-of-word and tw-idf: New approach to ad hoc ir. In Proceedings of the 22Nd ACM international conference on information and knowledge management, CIKM’13 (pp. 59–68). New York, NY, USA, ACM.
Rubin, D. L., Shah, N. H., & Noy, N. F. (2008). Biomedical ontologies: A functional perspective. Briefings in Bioinformatics, 9(1), 75–90.
Salton, G., & Buckley, C. (1988). Term-weighting approaches in automatic text retrieval. Information processing and management, 24(5), 513–523.
Singhal, A., Buckley, C., & Mitra, M. (1996). Pivoted document length normalization. In Proceedings of the 19th international ACM SIGIR conference on research and development in information retrieval, SIGIR’96 (pp. 21–29). New York, NY, USA, ACM.
Spasic, I., Greenwood, M., Preece, A., Francis, N., & Elwyn, G. (2013). FlexiTerm: a flexible term recognition method. Biomedical Semantics, 4(1), 27.
Stoykova, V., & Petkova, E. (2012). Automatic extraction of mathematical terms for precalculus. Procedia Technology Journal, 1, 464–468.
Tamura, A., Watanabe, T., & Sumita, E. (2012). Bilingual lexicon extraction from comparable corpora using label propagation. In Proceedings of the 2012 joint conference on empirical methods in natural language processing and computational natural language learning, EMNLP-CoNLL’12 (pp. 24–36). Stroudsburg, PA, USA, Association for Computational Linguistics.
Tian, Y., & Lo, D. (2015). A comparative study on the effectiveness of part-of-speech tagging techniques on bug reports. In Proceedings of the 22nd international IEEE conference on software analysis, evolution, and reengineering, SANER’15 (pp. 570–574). Montreal, Canada, IEEE.
Van Eck, N. J., Waltman, L., Noyons, E. C., & Buter, R. K. (2010). Automatic term identification for bibliometric mapping. Scientometrics, 82(3), 581–596.
Yang, Y., Zhao, T., Lu, Q., Zheng, D., & Yu, H. (2009). Chinese term extraction using different types of relevance. In Proceedings of the international joint conference on natural language processing, ACL-IJCNLP’09 (pp. 213–216). Suntec, Singapore, Association for Computational Linguistics.
Zadeh, R. B., & Goel, A. (2013). Dimension independent similarity computation. Journal of Machine Learning Research, 14(1), 1605–1626.
Zhang, X., Song, Y., & Fang, A. (2010). Term recognition using conditional random fields. In International conference on natural language processing and knowledge engineering, NLP-KE’10 (pp. 1–6). IEEE.
Zhang, Z., Iria, J., Brewster, C., & Ciravegna, F. (2008). A comparative evaluation of term recognition algorithms. In Proceedings of the sixth international conference on language resources and evaluation, LREC’08, Marrakech, Morocco.
Acknowledgments
This work was supported in part by the French National Research Agency under the JCJC program, Grant ANR-12-JS02-01001, as well as by the University of Montpellier, CNRS, IBC of Montpellier project and the FINCyT program, Peru. Finally, the authors thank DIST (Scientific and Technical Information Service) for the acquisition of Cirad corpus.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lossio-Ventura, J.A., Jonquet, C., Roche, M. et al. Biomedical term extraction: overview and a new methodology. Inf Retrieval J 19, 59–99 (2016). https://doi.org/10.1007/s10791-015-9262-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10791-015-9262-2