Abstract
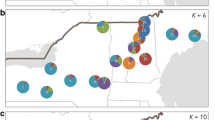
The abundance of butternut (Juglans cinerea L.) trees has severely declined rangewide over the past 50 years. An important factor in the decline is butternut canker, a disease caused by the fungus Ophiognomonia clavigigenti-juglandacearum, which has left the remaining butternuts isolated and sparsely distributed. To manage the remaining populations effectively, information regarding how butternut’s population genetic structure is affected by environmental and historical factors is needed. In this study, we assessed genetic structure and diversity of 161 butternut trees from 19 adjacent watersheds in the southern portion of butternut’s range using 12 microsatellite markers. We assessed the genetic diversity and genetic differentiation among trees grouped at various spatial scales. Our goal was to use historical abundance and land use data for these watersheds, which are now all a part of the Great Smoky Mountains National Park (GSMNP), to understand the ecological and evolutionary forces that challenge the conservation and management of butternut. In general, butternuts within the 19 neighboring watersheds were all part of one continuous population, with gene flow throughout. Significant genetic differentiation was detected between some groups of trees, but the differentiation was quite small and may not represent an ecologically significant distinction. The mean heterozygosity in all watersheds remained high, despite extensive mortality. Overall, genetic diversity and rare alleles were evenly distributed across all watersheds, with some variability in subpopulations containing butternut-Japanese walnut hybrids (Juglans x bixbyi or buarts). These results indicate that management of this species should focus on protection from future hybridization with Japanese walnut, promotion of regeneration, and persistence of all remaining butternut trees, which still retain high levels of genetic diversity.



Similar content being viewed by others
References
Altizer S, Harvell D, Friedle E (2003) Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol Evol 18:589–596
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10:556–557
Dutech C, Sork VL, Irwin AJ, Smouse PE, Davis FW (2005) Gene flow and fine-scale genetic structure in a wind-pollinated tree species, Quercus lobata (Fagaceaee). Am J Bot 92(2):252–261
Eckert AJ, Eckert ML, Hall BD (2010) Effects of historical demography and ecological context on spatial patterns of genetic diversity within foxtail pine (Pinus balfouriana; Pinaceae) stands located in the Klamath Mountains, California. Am J Bot 97:650–659
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–242
Epperson BK (1992) Spatial structure of genetic variation within populations of forest trees. New Forest 6:257–278
Epperson BK, Chung MG (2001) Spatial genetic structure of allozyme polymorphisms within populations of Pinus strobus (Pinaceae). Am J Bot 88:1006–1010
ESRI (Environmental Systems Resource Institute) (2009) ArcMap 9.3.1. ESRI, Redlands, California
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics 164:1567–1587
Goudet J (1995) FSTAT: a computer program to calculate F statistics, version 2.9.3. J Hered 86:485–486
Gugerli F, Walser JC, Dounavi K, Holderegger R, Finkeldey R (2007) Coincidence of small-scale spatial discontinuities in leaf morphology and nuclear microsatellite variation of Quercus petraea and Q. robur in a mixed forest. Ann Bot 99:713–722
Guillot G, Estoup A, Mortier F, Cosson JF (2005a) A spatial statistical model for landscape genetics. Genetics 170:1261–1280
Guillot G, Mortier F, Estoup A (2005b) Geneland: a computer package for landscape genetics. Mol Ecol Resour 5:712–715
Hamrick JL (1982) Plant population genetics and evolution. Am J Bot 69:1685–1693
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Vekemans X (2007) SPAGeDi 1.2—a program for spatial pattern analysis of genetic diversity. User’s manual. Available from http://www.ulb.ac.be/sciences/ecoevol/spagedi.html
Hoban SM, Anderson R, McCleary T, Schlarbaum S, Romero-Severson J (2008) Thirteen nuclear microsatellite loci for butternut (Juglans cinerea L.). Mol Ecol Resour 8:643–646
Hoban SM, McCleary TS, Schlarbaum SE, Romero-Severson J (2009) Geographically extensive hybridization between the forest trees American butternut and Japanese walnut. Biol Lett 5:324–327
Hoban SM, Borkowski DS, Brosi SL, McCleary TS, Thompson LM, McLachlan JS, Pereira MA, Schlarbaum SE, Romero-Severson J (2010) Range-wide distribution of genetic diversity in the North American tree Juglans cinerea: a product of range shifts, not ecological marginality or recent population decline. Mol Ecol 19:4876–4891
Hoban SM, Schlarbaum SE, Brosi SL, Romero-Severson J (2012a) A rare case of natural regeneration in butternut, a threatened forest tree, is parent and space limited. Conserv Genet 13:1447–1457
Hoban SM, McCleary TS, Schlarbaum SE, Anagnostakis SL, Romero-Severson J (2012b) Human‐impacted landscapes facilitate hybridization between a native and an introduced tree. Evol Appl 5(7):720–731
Holderegger R, Wagner HH (2008) Landscape genetics. Bioscience 58:199–207
Holderegger R, Buehler D, Gugerli F, Manel S (2010) Landscape genetics of plants. Trends Plant Sci 15:675–683
Jenkins MA (2007) Vegetation communities of the Great Smoky Mountains National Park. Southeast Nat 6(Special Issue 1):35–56
Jenkins MA, White PS (2002) Cornus florida L. mortality and understory composition changes in western Great Smoky Mountains National Park. J Torrey Bot Soc 129:194–206
Jones FA, Hamrick JL, Peterson CJ, Squiers ER (2006) Inferring colonization history from analyses of spatial genetic structure within populations of Pinus strobus and Quercus rubra. Mol Ecol 15:851–861
Jost L (2008) G ST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Kalisz S, Nason JD, Hanzawa FM, Tonsor SJ (2001) Spatial population genetic structure of Trillium grandiflorum: the roles of dispersal, mating, history and selection. Evolution 55:1560–1568
Kitamoto N, Honjo M, Ueno S, Takenaka A, Tsumura Y, Washitani I, Ohsawa R (2005) Spatial genetic structure among and within populations of Primula sieboldii growing beside separate streams. Mol Ecol 14:149–157
Knowles P, Perry DJ, Foster H (1992) Spatial genetic structure in two tamarack [Larix laricina (Du ROI) K. Koch] populations with differing establishment histories. Evolution 46:572–576
Levin DA, Franscisco-Oretega J, Jansen RK (1996) Hybridization and extinction of rare plant species. Conserv Biol 10:10–16
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Syst 15:65–95
Mccleary TS, Robichaud RL, Nuanes S, Anagnostakis SL, Schlarbaum SE, Romero-Severson J (2009) Four cleaved amplified polymorphic sequence (CAPS) markers for the detection of the Juglans ailantifolia chloroplast in putatively native J. cinerea populations. Mol Ecol Resour 9:525–527
Michler CH, Pijut PM, Jacobs DF, Meilan R, Woeste KE, Ostry ME (2006) Improving disease resistance of butternut (Juglans cinerea), a threatened fine hardwood: a case for single-tree selection through genetic improvement and deployment. Tree Physiol 26:121–128
Nei M (1978) Estimation of average heterozygosity and genetic distance for a small number of individuals. Genetics 89:583–590
Ostry ME, Woeste K (2004) Spread of butternut canker in North America, host range, evidence of resistance within butternut populations and conservation genetics. In: Michler CH, Pijut PM, van Sambeek J, Coggeshall M, Seifert J, Woeste K, Overton R (eds) Black walnut in a new century: Proc. 6th Walnut Council Research Symposium.. USDA For Serv, North Central Res Stn, Gen Tech Rep NC-243, St. Paul, MN, pp 114–120
Ouborg NJ, Francesco A, Vergeer P (2010) An essay on the necessity and feasibility of conservation genomics. Conserv Genet 11:643–653
Parker CR, Pipes DW (1990) Watersheds of Great Smoky Mountains National Park: a geographical information system analysis. Uplands Field Research Laboratory, Great Smoky Mountains National Park
Parks AM, Jenkins MA, Woeste KE, Ostry ME (2013) Conservation status of a threatened tree species: establishing a baseline for restoration of Juglans cinerea L. in the southern Appalachian Mountains. Natural Areas Journal (in press)
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Petit RJ, Abdelhamid EM, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Pons O, Petit RJ (1995) Estimation, variance and optimal sampling of gene diversity. Theor Appl Genet 90:462–470
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pyle C (1985) Vegetation disturbance history of Great Smoky Mountains National Park: an analysis of archival maps and records. USDI, NPS, Res/Resour Manage Rep SER-77, Southeast Regional Office, Atlanta, Georgia, 69 p
Pyle C (1988) The type and extent of anthropogenic vegetation disturbance in the great Smoky Mountains before National Park Service acquisition. Castanea 53:183–196
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27:83–109
Rink G (1990) Juglans cinerea L., butternut. In: Burns RM, Honkala BH (eds) Silvics of North America. Vol. 2. Hardwoods. Tech. Coords. . USDA Forest Service Agricultural Handbook 654, Washington, DC, pp 386–390
Rosenberg MS, Anderson CD (2011) PASSaGE: pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol Evol 2:229–232
Ross-Davis A, Woeste K (2008) Microsatellite markers for Juglans cinerea L. and their utility in other Juglandaceae species. Conserv Genet 9:465–469
Ross-Davis A, Huang Z, McKenna J, Ostry M, Woeste K (2008a) Morphological and molecular methods to identify butternut (Juglans cinerea) and butternut hybrids: relevance to butternut conservation. Tree Physiol 28:1127–1133
Ross-Davis A, Ostry M, Woeste K (2008b) Genetic diversity of butternut (Juglans cinerea) and implications for conservation. Can J For Res 38:899–907
Schultz J (2003) Conservation assessment for butternut or white walnut (Juglans cinerea L.). USDA For Serv, Eastern Region. Milwaukee, WI, 76 p
Segelbacher G, Cushman SA, Epperson BK, Fortin M, Francois O, Hardy OJ, Holderegger R, Taberlet P, Waits LP, Manel S (2010) Applications of landscape genetics in conservation biology: concepts and challenges. Conserv Genet 11:375–385
Sork VL, Smouse PE (2006) Genetic analysis of landscape connectivity in tree populations. Landscape Ecol 21:821–836
Sork VL, Waits L (2010) Contributions of landscape genetics—approaches, insights, and future potential. Mol Ecol 19:3489–3495
Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP (2010) Landscape genetics: where are we now? Mol Ecol 19:3496–3514
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Victory ER, Glaubitz JC, Rhodes OE Jr, Woeste K (2006) Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am J Bot 93(1):118–126
Victory ER, Glaubitz JC, Fike JA, Rhodes OE Jr., Woeste K (2008) Different histories but similar genetic diversity and structure for black walnut in Indiana and Missouri. In: Jacobs DF, Michler CH. Proceedings, 16th central hardwood forest conference. Gen. Tech. Rep. NRS-P-24. Newtown Square, PA: USDA For Serv, Northern Res Stn, pp 436-445
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Woeste K, Farlee L, Ostry M, McKenna J, Weeks S (2009) A forest manager’s guide to butternut. North J Appl For 26:9–14
Zhao P, Woeste KE (2010) DNA markers identify hybrids between butternut (Juglans cinerea L.) and Japanese walnut (Juglans ailantifolia Carr.). Tree Genet Genomes 7:511–533
Acknowledgments
We would like to thank the employees and volunteers of Great Smoky Mountains National Park for providing invaluable support for this project, especially Tom Remaley, Janet Rock, Keith Langdon, Kristine Johnson, Glenn Taylor, Emily Darling, Emily Guss, and Paul Super for contributing their time and resources to facilitate our work in GSMNP. Field assistance was provided by H. Seiner, B. Greene, V. Meyer, B. Moehl, J. Albritton, M. Cooke, S. Simpson, J. Byrne, M. Duffy, M. Clebsch, C. Pan, M. Williams, K. Hansen, C. deGuia, L. Combs, C. Black, D. Love, D. Clabo, and K. Mann. William Beatty provided useful advice for statistical analyses. This work was supported by the Hardwood Tree Improvement and Regeneration Center (United States Department of Agriculture Forest Service Northern Research Station) and the Department of Forestry and Natural Resources at Purdue University. Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the US Department of Agriculture and does not imply its approval to the exclusion of other products or vendors that also may be suitable.
Data archiving statement
Microsatellite genotypes at all loci for all samples including hybrids, and the watershed from which the samples were obtained, are archived at TreeGenes as TGDR007. The specific locations of sampled trees are available for research purposes from Great Smoky Mountains National Park; contact Tom Remaley, Supervisory Ecologist/Inventory & Monitoring Coordinator, TOM_REMALEY@NPS.GOV.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. G. Vendramin
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1
(DOCX 51 kb)
Rights and permissions
About this article
Cite this article
Parks, A., Jenkins, M., Ostry, M. et al. Biotic and abiotic factors affecting the genetic structure and diversity of butternut in the southern Appalachian Mountains, USA. Tree Genetics & Genomes 10, 541–554 (2014). https://doi.org/10.1007/s11295-014-0702-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0702-8