Abstract
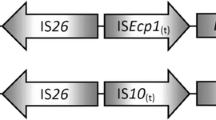
Untreated wastewater is a risk factor for the spread of antibiotic resistance in the environment. However, little is known about the contribution of untreated wastewater to the burden of antibiotic resistance in the Nigerian environment. In this study, a total of 143 ceftazidime-/cefpodoxime-resistant bacteria isolated from untreated wastewater and untreated wastewater-contaminated surface and groundwater in Nigeria were screened for extended-spectrum β-lactamase (ESBL) genes, integrons and integron gene cassettes by PCR. The genetic environment of bla CTX-M-15 was mapped by PCR and potentially conjugative plasmids were detected among the isolates by degenerate primer MOB typing (DPMT). ESBL production was confirmed in 114 (79.7%) isolates and ESBL genes (bla SHV, bla CTX-M-15 and bla TEM) were detected in 85 (74.6%) ESBL-producing isolates. bla CTX-M-15 was associated with ISEcp1 and with orf477 in 12 isolates and with ISEcp1, IS26 and orf477 in six others. To the best of our knowledge, this is the first report of bla CTX-M-15 in hand-dug wells and borehole serving as sources of drinking water and a first report of the genetic environment of bla CTX-M-15 in environmental bacteria from Nigeria. The results of this study confirm untreated wastewater as an important medium for the spread of ESBL-producing bacteria within the Nigerian environment. Hence, the widespread practice of discharging untreated wastewater into the aquatic ecosystem in Nigeria is a serious risk to public health.

Similar content being viewed by others
References
Abgottspon H, Nuesch-Inderbinen MT, Zurfluh K et al (2014) Enterobacteriaceae with extended-spectrum- and pAmpC-type betalactamase-encoding genes isolated from freshwater fish from two lakes in Switzerland. Antimicrob Agents Chemother 58:2482–2484
Aibinu I, Pfeifer Y, Peters F, Ogunsola F, Adeonipekun E, Odungbemi T, Koenig W (2012) Emergence of bla CTX-M-15, qnrB1 and acc(6′)-Ib-cr resistance genes in Pantoea agglomerans and Enterobacter cloacae from Nigeria (sub-Saharan Africa). J Med Microbiol 61:165–167
Alcala L, Alonso CA, Simon C, Gonzalez-Esteban C, Oros J, Rezusta A et al (2016) Wild birds, frequent carriers of extended-Spectrum beta-lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb Ecol 72:861–869
Alvarado A, Garcillan-Barcia MP, de la Cruz F (2012) A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One 7(7):e40438. https://doi.org/10.1371/journal.pone.0040438
Amos GCA, Hawkey PM, Gaze WH et al (2014) Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J Antimicrob Chemother 69:1785–1791
Berendonk TU, Manaia CM, Merlin C et al (2015) Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13:310–317
Brooke JS (2012) Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41
Buck JD (1982) Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Calbo E, Freixas N, Xercavins M et al (2011) Foodborne nosocomial outbreak of SHV1 and CTX-M-15-producing Klebsiella pneumonia: epidemiology and control. Clin Infect Dis 52:743–749
Canton R (2009) Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin Microbiol Infect 15(Suppl.I):20–25
Clinical and Laboratory Standards Institute (CLSI) (2011) Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement M100-S21. CLSI, Wayne
Czekalski N, Gascon-Díez E, Bürgmann H (2014) Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J 8:1381e1390
Di Cesare A, Eckert EM, D'Urso S, Bertoni R, Gillan DC, Wattiez R, Corno G (2016) Co-occurrence of integrase 1, antibiotic and metal resistance genes in municipal wastewater treatment plants. Water Res 94:208–214
Dihanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N (2011) Variation in the genetic environments of blaCTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66:1005–1012
Dolejska M, Frolkova P, Florek M, Jamborova I, Purgertova M, Kutilova I, Cizak A, Guenther S, Literak I (2011) CTX-M-15-producing Escherichia coli clone B2-025b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J Antimicrob Chemother 66:2784–2790
Du J, Ren H, Geng J, Zhang Y, Xu K, Ding L (2014) Occurrence and abundance of tetracycline, sulfonamide resistance genes, and class 1 integron in five wastewater treatment plants. Environ Sci Pollut R 21:7276–7284
Finley RL, Collignon P, Joakim-Larsson DG, McEwen SA, Li XZ, Gaze WH, Smith RR, Timinouni M, Graham DW, Topp E (2013) The scourge of antibiotic resistance and the important role of the environment. CID 57:704–710
Fortini D, Fashae K, Villa L, Feudi C, García-Fernández A, Carattoli A (2015) A novel plasmid carrying bla CTX-M-15 identified in commensal Escherichia coli from healthy pregnant women in Ibadan, Nigeria. J Global Antimicrob Resist 3:9–12
Huang YW, Hu RM, Lin YT et al (2014) Contribution of class 1 integron to antimicrobial resistance in Stenotrophomonas maltophilia. Microb Drug Resist. https://doi.org/10.1089/mdr.2014.0072
Iroha IR, Esimone CO, Neumann S, Marlinghaus L, Koerte M, Szabados F, Gatermann S, Kaase M (2012) First description of Escherichia coli producing CTX-M-15-extended spectrum beta lactamase (ESBL) in out-patients from south eastern Nigeria. Ann Clin Microbiol Antimicrob 11:19. https://doi.org/10.1186/1476-0711-11-19
Inwezerua C, Mendonça N, Calhau V, Domingues S, Adeleke OE, Da Silva GJ (2014) Occurrence of extended-spectrum beta-lactamases in human and bovine isolates of Escherichia coli from Oyo State, Nigeria. J Infect Dev Ctries 8(6):774–779
Kasap M, Fashae K, Torol S, Kolayli F, Budak F, Vahaboglu H (2010) Characterization of ESBL (SHV-12) producing clinical isolate of Enterobacter aerogenes from a tertiary care hospital in Nigeria. Ann Clin Microbiol Antimicrob 2010 9:1. http://www.ann-clinmicrob.com/content/9/1/1
Korzeniewska E, Harnisz M (2013) Extended spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. J Environ Manag 128:904–911
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrant E, Goodfellov M (eds) Nucleic acids techniques in bacterial systematics. Wiley, Chichester, pp 115–148
LaPara TM, Burch TR, McNamara PJ, Tan DT, Yan M, Eichmiller JJ (2011) Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ Sci Technol 45(22):9543–9549
Lupo A, Coyne S, Berendonk TU (2012) Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front Microbio 3:18. https://doi.org/10.3389/fmicb.2012.00018
Machado E, Canton R, Baquero F et al (2005) Integron content of extended-spectrum-b-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob Agents Chemother 49(5):1823–1829
Maravic A, Skocibusic M, Fredotovic Z et al (2014) Characterisation of environmental CTX-M-15 producing Stenotrophomonas maltophilia. Antimicrob Agents Chemother 58:6333–6334
Maravic A, Skocibusic M, Cvjetan S, Samanic I, Fredotovic Z, Puizina J (2015) Prevalence and diversity of extended-spectrum-beta-lactamase-producing Enterobacteriaceae from marine beach waters. Mar Pollut Bull 90:60–67
Munir M, Wong K, Xagoraraki I (2011) Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res 45:681–693
Mulvey MR, Soule G, Boyd D et al (2003) Characterisation of the first extended spectrum beta-lactamse producing Salmonella isolate in Canada. J Clin Microbiol 41:460–462
Novovic K, Filipic B, Veljovic K, Begovic J, Mirkovic N, Jovcic B (2015) Environmental waters and blaNDM-1 in Belgrade, Serbia: endemicity questioned. Sci Total Environ 511:393–398. https://doi.org/10.1016/j.scitotenv.2014.12.072
Obasi A, Nwachukwu SC, Ugoji E, Kholer C, Ghöler A, Balau V, Pfeifer Y, Steinmetz I (2017) Extended-spectrum β-lactamase-producing Klebsiella pneumoniae from pharmaceutical wastewaters in south-western Nigeria. Microb Drug Resist. https://doi.org/10.1089/mdr.2016.0269
Ogbolu DO, Daini OA, Ogunledun A, Oyebode A, Terry-Alli OA, Webber MA (2013) Dissemination of IncF plasmids carrying beta-lactamase genes in gram negative bacteria from Nigerian hospitals. J Infect Dev Ctries 7(5):382–390. https://doi.org/10.3855/jidc.2613
Ogbolu DO, Daini OA, Ogunledun A, Alli AO, Webber MA (2011) High levels of multidrug resistance in clinical isolates of Gram-negative pathogens from Nigeria. Int J Antimicrob Agents 37:62–66
Ojo OE, Schwarz S, Michael GB (2016) Detection and characterization of extended-spectrum-β-lactamase-producing Escherichia coli from chicken production chains in Nigeria. Vet Microbiol 194:62–68
Orsini M, Romano-Spica V (2001) A microwave-based method for nucleic acid isolation from environmental samples. Lett Appl Microbiol 33:17–20
Ozaktas T, Taskin B, Gozen AG (2012) High level multiple antibiotic resistance among fish surface associated bacterial populations in non-aquaculture freshwater environment. Water Res 46:6382–6390. https://doi.org/10.1016/j.watres.2012.09.010
Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162
Pouget JG, Coutinho FJ, Reid-Smith RJ, Boerlin P (2013) Characterization of blaSHV genes on plasmids from Escherichia coli and Salmonella enterica isolates from Canadian food animals (2006-2007). Appl Environ Microbiol 79:3864–3866
Raji MA, Jamal W, Ojemeh O, Rotimi VO (2015) Sequence analysis of genes mediating extended-spectrum beta-lactamase (ESBL) production in isolates of Enterobacteriaceae in a Lagos Teaching Hospital, Nigeria. BMC Infect Dis 7:215–259
Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sànchez-Melsió A, Borrego CM, Barceló D, Balcázar JL (2015) Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res 69:234–242
Rocha-Gracia RC, Cortés-Cortés G, Lozano-Zarain P, Bello F, Martínez-Laguna Y, Torres C (2015) Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet J 203:315–319
Soge OO, Adeniyi BA, Roberts MC (2006) New antibiotic resistance genes associated with CTX-M plasmids from uropathogenic Nigerian Klebsiella pneumoniae. J Antimicrob Chemother 58:1048–1053
Tacão M, Correia A, Henriques I (2012) Resistance to broadspectrum antibiotics in aquatic systems: anthropogenic activities modulate the dissemination of bla CTX-M-like genes. Appl Environ Microbiol 78:4134–4140
Tada T, Miyoshi-Akiyama T, Dahal RK et al (2014) Identification of a novel 6′-N-aminoglycoside aminotransferase, ACC(6′)-lak, from a multidrug resistant clinical isolate of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 58:6324–6327
Toleman MA, Bennett PM, Bennett DMC et al (2007) Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by the acquisition of sul genes. Emerg Infect Dis 13:559–565
Zurfluh K, Hachler H, Nuesch-Inderbinen M, Stephan R (2013) Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microbiol 79:3021–3026
Zurfluh K, Nüesch-Inderbinen M, Morach M, Zihler Berner A, Hächler H, Stephan R (2015) Extended-spectrum-β-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl Environ Microbiol 81:3115–3120
Acknowledgements
We thank Dr. Dayo Sowunmi of the Department of Zoology, University of Ibadan, Nigeria, for his invaluable assistance in sample collection from Ikpoba and Cross Rivers and Dr. Jochen Müller of the Department of Environmental Biotechnology, Helmholtz Center for Environmental Research, Leipzig, Germany, for his assistance in the course of this project.
Funding
This work was supported by the ANTI-resist (02WRS1272A) Bundesministerium für Bildung und Forschung (BMBF), Germany risk management of new pollutants and pathogens in the water cycle (Riskwa) and the JPI water STARE project. O.O.A. was supported by the German Academic Exchange Service (DAAD) under the University Academics and Scientists Fellowship program during his stay at the Technical University of Dresden.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Diane Purchase
Rights and permissions
About this article
Cite this article
Adelowo, O.O., Caucci, S., Banjo, O.A. et al. Extended Spectrum Beta-Lactamase (ESBL)-producing bacteria isolated from hospital wastewaters, rivers and aquaculture sources in Nigeria. Environ Sci Pollut Res 25, 2744–2755 (2018). https://doi.org/10.1007/s11356-017-0686-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-0686-7