-
PDF
- Split View
-
Views
-
Cite
Cite
Fabrice Merien, Denis Portnoi, Pascale Bourhy, Françoise Charavay, Alain Berlioz-Arthaud, Guy Baranton, A rapid and quantitative method for the detection of Leptospira species in human leptospirosis, FEMS Microbiology Letters, Volume 249, Issue 1, August 2005, Pages 139–147, https://doi.org/10.1016/j.femsle.2005.06.011
Close - Share Icon Share
Abstract
Prompt laboratory diagnosis of leptospirosis infection facilitates patient management and initiation of therapy. A cost effective real-time PCR assay using SYBR Green I was developed for detection of pathogenic leptospires in serum specimens. Specific PCR products were obtained only with DNA of pathogenic Leptospira genomospecies. LightCycler PCR ability to distinguish between species was possible using melting curves, providing an approach for identification with a specific Tm assigned to a single species or set of species. Assay sensitivity was approximately 50 leptospires/ml, corresponding to one to two genome copies in a PCR mixture. Fifty-one patients who had clinical symptoms consistent with leptospirosis were tested both with a previously described rrs amplification and our real-time assay. Our LFB1 real-time assay confirmed the diagnosis for 25 patients (49%, 25/51) and revealed an estimated density of 8.0 × 101–3.9 × 104 leptospires/ml of blood. The total assay time for 12 clinical samples from sample to data analysis was less than 3 h. These data illustrate the potential of our LFB1 real-time assay for the rapid detection of leptospires in serum samples and their subsequent quantification in a single run.
1 Introduction
Leptospirosis is a zoonotic infection with a worldwide distribution affecting wild and domestic vertebrates, the incidence of the disease being higher in tropical climates [[]. This disease remains underdiagnosed largely due to the broad spectrum of signs and symptoms attributable to this spirochetal pathogen [[]. Severe cases are not infrequent and early antibiotic therapy is critical for their management since a high bacterial load in the blood is a bad prognosis [[]. The causative bacteria, pathogenic Leptospira species, are transmitted to human most commonly by indirect contact with contaminated fresh water. Although classification based on serotyping remains the gold standard, molecular-based taxonomy, which is still in use, classifies Leptospira into 12 genomically distinct species: Leptospira alexanderi, Leptospira biflexa, Leptospira borgpetersenii, Leptospira fainei, Leptospira inadai, Leptospira interrogans, Leptospira kirschneri, Leptospira noguchii, Leptospira santarosai, Leptospira weilii, Leptospira meyeri and Leptospira wolbachii[[,[]. This molecular-based classification still co-exists with serotyping for which there is no sufficiently valid argument so that the most recent system may eventually replace the oldest.
Conventional laboratory diagnosis usually depends on culture and serological techniques like microscopic agglutination test (MAT), ELISA and dipstick assay [[]. Briefly, biological confirmation of leptospirosis is laborious and does not provide a rapid diagnosis. Isolation of leptospires from human blood is possible in the acute phase of the disease that lasts for up to about 10 days. Culture in EMJH (Ellinghausen McCullough Johnson and Harris) medium may take up to 2 months and does not provide an emergency diagnosis. Serological techniques are mainly used; however, antibodies are undetectable before 8–10 days after disease onset. Moreover, MAT requires the maintenance of a large number of live Leptospira strains as source of antigens and paired serum samples are needed for the correct interpretation of the results [[].
The polymerase chain reaction (PCR) has the potential to make a dramatic impact in diagnosing leptospirosis. Historically, several pairs of primers for PCR detection of leptospires have been published, some based on specific gene targets (rRNA genes), repetitive elements or sequences derived from genomic libraries [[]. In the early stage of an outbreak, it is important to confirm that DNA sequences that have been amplified are definitively derived from Leptospira. This can be done by hybridization [[], nested-PCR or direct sequencing of amplicons [[]. Each of these approaches, however, requires additional manipulations and thus adds time to the diagnostic procedure. In our first assay [[], a combined PCR-hybridization test (digoxygenin-labelled probe) was proposed for qualitative detection of a specific target sequence of Leptospira. The primers amplified a 331-bp fragment of the rrs (16S rRNA gene) of both pathogenic and non-pathogenic leptospires. A minimum of 36–48 h was necessary to give a result to the physician and evaluation of bacterial density was not possible. Then, a nested-PCR with internal primers was introduced in the initial protocol to replace the overnight hybridization step and shorten the diagnostic method. However, as a major issue with nested PCR, carry-over contamination was always possible (unpublished data). In order to follow the course of acute human leptospirosis, an ELISA microtiter plate hybridization method was developed for quantitative determination of Leptospira spp. in biological samples following PCR [[]. We obtained evidence that a density of 104 leptospires/ml of blood is a critical threshold for the vital prognosis of the patients [[]. The practical nature of this method makes it suitable for a limited number of samples but not for routine diagnosis with high throughput testing. This quantitative PCR assay was used to monitor the density of leptospires in blood and in target organs in a Syrian hamster model and to determine the efficacy of antibiotics (ampicillin, deoxycycline and ofloxacin) during the course of the disease [[]. These findings emphasize the need for highly sensitive and quantitative assays for clinical studies.
In this work, we report the development of a quantitative real-time PCR assay using the SYBR Green detection technology that represents a new, rapid, sensitive and efficient tool for molecular detection and identification of pathogenic leptospires in clinical specimens.
2 Materials and methods
2.1 DNA extraction for rrs amplification
DNA was purified from 100 μl of serum using silica particles and guanidium thiocyanate according to the method of Boom et al. [[]. DNA was eluted in 50 μl of TE buffer and 5 μl was used as template in the first PCR of the nested assay.
2.2 Nested-PCR for rrs amplification
A 331-bp fragment was amplified in a total volume of 50 μl using a pair of primers designed from the 16S rRNA gene of Leptospira species, as previously described [[]. Then, a second amplification in a total volume of 50 μl was performed with internal primers C (CAAGTCAAGCGGAGTAGCAA) and D (CTTAACTGCTGCCTCCCGTA). The reaction mixture consisted of Taq 10× reaction buffer (Promega, USA), 2 mM MgCl2, 1 μM of each oligonucleotide primer (Proligo Singapore Pte Ltd), 200 μM dNTPs (Roche Applied Science, New Zealand) and 1.25 U of Red Hot DNA polymerase (Fisher Scientific, USA). Five μl of a 1 in 1000 dilution, using PCR grade water, of the first amplification was then added to 45 μl of the reaction mixture. PCR was performed in a Perkin Elmer Cetus 9600 thermal cycler with 20 cycles consisting of denaturation at 94°C for 30 s, annealing at 61°C for 1 min, and extension at 72°C for 1 min. The last cycle consisted of denaturation at 94°C for 1 min, annealing at 61°C for 1.5 min, and extension at 72°C for 10 min. Then the final 289-bp amplification product was analysed by gel electrophoresis in a 2% NuSieve 3:1 gel agarose (FMC BioProducts, Rockland, ME) with ethidium bromide staining. This high gel strength results in easy-to-handle gels, enhancing the convenience of gel processing and ensuring fine resolution of DNA fragments up to 1000 bp.
2.3 DNA extraction for real-time PCR
Total DNA from human serum (200 μl) was isolated by using the QIAamp DNA Mini Kit (Qiagen, Australia). Serum specimens were processed as follows: 200 μl of lysis buffer and 20 μl of proteinase K solution were mixed with 200 μl of serum and incubated at 56°C for 30 min in a water bath. Wash steps were performed according to the manufacturer's instructions by using AW1 and AW2 buffers. DNA was eluted with 50 μl of AE buffer. In selected cases, urine specimens were tested. Briefly, 1.2 ml of urine was centrifuged at 12,000g for 20 min, and the pellet was suspended in 200 μl of PCR grade water (Roche Applied Science, New Zealand) and vortexed vigorously. Then, DNA was extracted under the same conditions of time and temperature as reported above. Eluted DNAs were stored at 20°C until use. Randomly selected negative Leptospira spp. culture samples were used as negative controls. Aerosol-barrier pipette tips were used throughout the procedure.
2.4 Design of real-time PCR primers
Primer LFB1-F (5′-CATTCATGTTTCGAATCATTTCAAA-3′) and primer LFB1-R (5′-GGCCCAAGTTCCTTCTAAAAG-3′) chosen in the locus LA0322 of L. interrogans Lai sequence [[0] were used to amplify a 331-bp product from pathogenic Leptospira spp. Primers were synthesized by Proligo Singapore Pte Ltd.
2.5 Real-time PCR with SYBR Green I
The LightCycler FastStart DNA Master SYBR Green I kit (Roche Applied Science, New Zealand) was used as the basis for the reaction mixture, using a 20 μl volume in each reaction capillary. The final reaction mix included a dNTP mix (with dUTP instead of dTTP), SYBR Green I dye, the hot-start enzyme FastStart Taq DNA polymerase, 5 mM MgCl2 and 0.5 μM of each primer. After distributing 15 μl aliquots of the mix to each capillary, 5 μl of the eluted DNA was then added before being capped, centrifuged and placed in the LightCycler sample carousel. A negative control with PCR-grade water rather than template DNA was always used with the samples. Aerosol-barrier pipette tips were used throughout the procedure. Cycle conditions were optimized regarding annealing temperature and holding times. Amplification conditions involved a pre-incubation at 95°C for 10 min (FastStart Taq DNA polymerase activation) followed by amplification of the target DNA for 50 cycles (95°C for 8 s, 60°C for 5 s and 72°C for 12 s) with a transition rate of 20°C/s. Melting curve analysis was performed at a linear temperature transition rate of 0.1°C/s from 65 to 95°C with continuous fluorescence acquisition. This step was followed by a cooling step at 40°C for 30 s. Performed automatically by the software, the first derivative of the initial melting curve (−dF/dT) was plotted against temperature for improved determination of the melting temperature (Tm). All experiments were repeated at least twice for reproducibility.
2.6 Reconstitution experiments with biological samples
Serum specimens from patients with diseases other than leptospirosis were seeded with leptospires (L. interrogans strain Verdun) and used for PCR. Leptospires were serially diluted from 5 × 104 to 50 bacteria/ml of serum. Artificially seeded urine was also prepared under the same conditions. DNA from all reconstituted samples was extracted as described for the real-time PCR.
2.7 Leptospira strains
Cultured leptospires from the Institut Pasteur collection were tested by PCR. Representatives of non-pathogenic and pathogenic Leptospira species were included in the study (Table 1). All strains were cultured in EMJH (Ellinghausen McCullough Johnson and Harris) medium and grown for up to seven days at 30°C to stationary phase to a density of approximately 108 cells/ml. Then, Leptospira DNA was extracted and purified as described by Brenner et al. [[1].
List of leptospiral strains used for nested rrs and LFB1 SYBRGreen PCR assays and results after amplification
| Genomospecies | Serovar | Strain | Nested rrs | LFB1 PCR | |
| L. interrogans | s.s. | icterohaemorrhagiae | Lai | + | + |
| L. interrogans | s.s. | icterohaemorrhagiae | Verdun | + | + |
| L. interrogans | s.s. | nicterohaemorrhagiae | RGA | + | + |
| L. interrogans | s.s. | australis | Ballico | + | + |
| L. interrogans | s.s. | autumnalis | Akiyami A | + | + |
| L. interrogans | s.s. | bataviae | Van Tienen | + | + |
| L. interrogans | s.s. | canicola | Hond Utrecht IV | + | + |
| L. interrogans | s.s. | hebdomadis | Hebdomadis | + | + |
| L. interrogans | s.s. | pomona | Pomona | + | + |
| L. interrogans | s.s. | pyrogenes | Salinem | + | + |
| L. kirschneri | cynopter | 3522C | + | + | |
| L. kirschneri | grippotyphosa | Moskva V | + | + | |
| L. borgpetersenii | castellonis | Castellon 3 | + | + | |
| L. borgpetersenii | sejroe | M84 | + | + | |
| L. borgpetersenii | tarassovi | Mitis Johnson | + | + | |
| L. noguchii | panama | CZ 214K | + | + | |
| L. alexanderi | java | Mengla | + | + | |
| L. weilii | sarmin | Sarmin | + | + | |
| L. santarosai | hebdo | Borincana | + | + | |
| L. inadai | lyme | Lyme 10 | + | − | |
| L. fainei | hurstbridge | BUT6 | + | − | |
| L. biflexa | s.s. | patoc | Patoc I | + | − |
| L. meyeri | semaranga | Veldrat | + | − | |
| L. wolbachi | codice | CDC | + | − |
| Genomospecies | Serovar | Strain | Nested rrs | LFB1 PCR | |
| L. interrogans | s.s. | icterohaemorrhagiae | Lai | + | + |
| L. interrogans | s.s. | icterohaemorrhagiae | Verdun | + | + |
| L. interrogans | s.s. | nicterohaemorrhagiae | RGA | + | + |
| L. interrogans | s.s. | australis | Ballico | + | + |
| L. interrogans | s.s. | autumnalis | Akiyami A | + | + |
| L. interrogans | s.s. | bataviae | Van Tienen | + | + |
| L. interrogans | s.s. | canicola | Hond Utrecht IV | + | + |
| L. interrogans | s.s. | hebdomadis | Hebdomadis | + | + |
| L. interrogans | s.s. | pomona | Pomona | + | + |
| L. interrogans | s.s. | pyrogenes | Salinem | + | + |
| L. kirschneri | cynopter | 3522C | + | + | |
| L. kirschneri | grippotyphosa | Moskva V | + | + | |
| L. borgpetersenii | castellonis | Castellon 3 | + | + | |
| L. borgpetersenii | sejroe | M84 | + | + | |
| L. borgpetersenii | tarassovi | Mitis Johnson | + | + | |
| L. noguchii | panama | CZ 214K | + | + | |
| L. alexanderi | java | Mengla | + | + | |
| L. weilii | sarmin | Sarmin | + | + | |
| L. santarosai | hebdo | Borincana | + | + | |
| L. inadai | lyme | Lyme 10 | + | − | |
| L. fainei | hurstbridge | BUT6 | + | − | |
| L. biflexa | s.s. | patoc | Patoc I | + | − |
| L. meyeri | semaranga | Veldrat | + | − | |
| L. wolbachi | codice | CDC | + | − |
List of leptospiral strains used for nested rrs and LFB1 SYBRGreen PCR assays and results after amplification
| Genomospecies | Serovar | Strain | Nested rrs | LFB1 PCR | |
| L. interrogans | s.s. | icterohaemorrhagiae | Lai | + | + |
| L. interrogans | s.s. | icterohaemorrhagiae | Verdun | + | + |
| L. interrogans | s.s. | nicterohaemorrhagiae | RGA | + | + |
| L. interrogans | s.s. | australis | Ballico | + | + |
| L. interrogans | s.s. | autumnalis | Akiyami A | + | + |
| L. interrogans | s.s. | bataviae | Van Tienen | + | + |
| L. interrogans | s.s. | canicola | Hond Utrecht IV | + | + |
| L. interrogans | s.s. | hebdomadis | Hebdomadis | + | + |
| L. interrogans | s.s. | pomona | Pomona | + | + |
| L. interrogans | s.s. | pyrogenes | Salinem | + | + |
| L. kirschneri | cynopter | 3522C | + | + | |
| L. kirschneri | grippotyphosa | Moskva V | + | + | |
| L. borgpetersenii | castellonis | Castellon 3 | + | + | |
| L. borgpetersenii | sejroe | M84 | + | + | |
| L. borgpetersenii | tarassovi | Mitis Johnson | + | + | |
| L. noguchii | panama | CZ 214K | + | + | |
| L. alexanderi | java | Mengla | + | + | |
| L. weilii | sarmin | Sarmin | + | + | |
| L. santarosai | hebdo | Borincana | + | + | |
| L. inadai | lyme | Lyme 10 | + | − | |
| L. fainei | hurstbridge | BUT6 | + | − | |
| L. biflexa | s.s. | patoc | Patoc I | + | − |
| L. meyeri | semaranga | Veldrat | + | − | |
| L. wolbachi | codice | CDC | + | − |
| Genomospecies | Serovar | Strain | Nested rrs | LFB1 PCR | |
| L. interrogans | s.s. | icterohaemorrhagiae | Lai | + | + |
| L. interrogans | s.s. | icterohaemorrhagiae | Verdun | + | + |
| L. interrogans | s.s. | nicterohaemorrhagiae | RGA | + | + |
| L. interrogans | s.s. | australis | Ballico | + | + |
| L. interrogans | s.s. | autumnalis | Akiyami A | + | + |
| L. interrogans | s.s. | bataviae | Van Tienen | + | + |
| L. interrogans | s.s. | canicola | Hond Utrecht IV | + | + |
| L. interrogans | s.s. | hebdomadis | Hebdomadis | + | + |
| L. interrogans | s.s. | pomona | Pomona | + | + |
| L. interrogans | s.s. | pyrogenes | Salinem | + | + |
| L. kirschneri | cynopter | 3522C | + | + | |
| L. kirschneri | grippotyphosa | Moskva V | + | + | |
| L. borgpetersenii | castellonis | Castellon 3 | + | + | |
| L. borgpetersenii | sejroe | M84 | + | + | |
| L. borgpetersenii | tarassovi | Mitis Johnson | + | + | |
| L. noguchii | panama | CZ 214K | + | + | |
| L. alexanderi | java | Mengla | + | + | |
| L. weilii | sarmin | Sarmin | + | + | |
| L. santarosai | hebdo | Borincana | + | + | |
| L. inadai | lyme | Lyme 10 | + | − | |
| L. fainei | hurstbridge | BUT6 | + | − | |
| L. biflexa | s.s. | patoc | Patoc I | + | − |
| L. meyeri | semaranga | Veldrat | + | − | |
| L. wolbachi | codice | CDC | + | − |
2.8 Clinically encountered bacterial species
Bacterial strains other than members of the genus Leptospira were subcultured (Table 2) and the respective DNAs were extracted and purified as described by Brenner et al. [[1].
List of non-leptospiral strains used for specificity assay
| Strain | Source |
| Acinetobacter baumanii | Laboratory isolate |
| Bacillus cereus | Laboratory isolate |
| Chryseobacterium meningosepticum | Laboratory isolate |
| Enterobacter hormaechei | Laboratory isolate |
| Enterococcus casseliflavus | Laboratory isolate |
| Enterococcus faecalis | Laboratory isolate |
| Escherichia coli | Laboratory isolate |
| Flavobacterium sp. | Laboratory isolate |
| Klebsiella pneumoniae | Laboratory isolate |
| Leclercia adecarboxylata | Laboratory isolate |
| Proteus mirabilis | Laboratory isolate |
| Salmonella enteritidis | Laboratory isolate |
| Shigella flexneri | Laboratory isolate |
| Staphylococcus aureus | Laboratory isolate |
| Staphylococcus epidermidis | Laboratory isolate |
| Staphylococcus pulvereri | Laboratory isolate |
| Staphylococcus succinis | Laboratory isolate |
| Stenotrophomonas maltophilia | Laboratory isolate |
| Strain | Source |
| Acinetobacter baumanii | Laboratory isolate |
| Bacillus cereus | Laboratory isolate |
| Chryseobacterium meningosepticum | Laboratory isolate |
| Enterobacter hormaechei | Laboratory isolate |
| Enterococcus casseliflavus | Laboratory isolate |
| Enterococcus faecalis | Laboratory isolate |
| Escherichia coli | Laboratory isolate |
| Flavobacterium sp. | Laboratory isolate |
| Klebsiella pneumoniae | Laboratory isolate |
| Leclercia adecarboxylata | Laboratory isolate |
| Proteus mirabilis | Laboratory isolate |
| Salmonella enteritidis | Laboratory isolate |
| Shigella flexneri | Laboratory isolate |
| Staphylococcus aureus | Laboratory isolate |
| Staphylococcus epidermidis | Laboratory isolate |
| Staphylococcus pulvereri | Laboratory isolate |
| Staphylococcus succinis | Laboratory isolate |
| Stenotrophomonas maltophilia | Laboratory isolate |
List of non-leptospiral strains used for specificity assay
| Strain | Source |
| Acinetobacter baumanii | Laboratory isolate |
| Bacillus cereus | Laboratory isolate |
| Chryseobacterium meningosepticum | Laboratory isolate |
| Enterobacter hormaechei | Laboratory isolate |
| Enterococcus casseliflavus | Laboratory isolate |
| Enterococcus faecalis | Laboratory isolate |
| Escherichia coli | Laboratory isolate |
| Flavobacterium sp. | Laboratory isolate |
| Klebsiella pneumoniae | Laboratory isolate |
| Leclercia adecarboxylata | Laboratory isolate |
| Proteus mirabilis | Laboratory isolate |
| Salmonella enteritidis | Laboratory isolate |
| Shigella flexneri | Laboratory isolate |
| Staphylococcus aureus | Laboratory isolate |
| Staphylococcus epidermidis | Laboratory isolate |
| Staphylococcus pulvereri | Laboratory isolate |
| Staphylococcus succinis | Laboratory isolate |
| Stenotrophomonas maltophilia | Laboratory isolate |
| Strain | Source |
| Acinetobacter baumanii | Laboratory isolate |
| Bacillus cereus | Laboratory isolate |
| Chryseobacterium meningosepticum | Laboratory isolate |
| Enterobacter hormaechei | Laboratory isolate |
| Enterococcus casseliflavus | Laboratory isolate |
| Enterococcus faecalis | Laboratory isolate |
| Escherichia coli | Laboratory isolate |
| Flavobacterium sp. | Laboratory isolate |
| Klebsiella pneumoniae | Laboratory isolate |
| Leclercia adecarboxylata | Laboratory isolate |
| Proteus mirabilis | Laboratory isolate |
| Salmonella enteritidis | Laboratory isolate |
| Shigella flexneri | Laboratory isolate |
| Staphylococcus aureus | Laboratory isolate |
| Staphylococcus epidermidis | Laboratory isolate |
| Staphylococcus pulvereri | Laboratory isolate |
| Staphylococcus succinis | Laboratory isolate |
| Stenotrophomonas maltophilia | Laboratory isolate |
3 Results
3.1 Specificity of the PCR assay
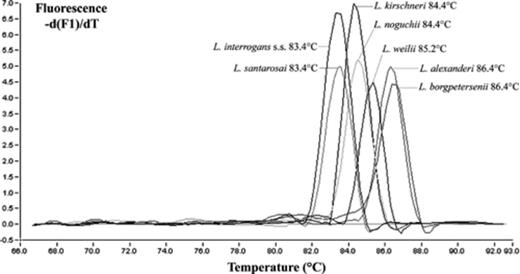
To assess the specificity of the primers, DNA samples of reference leptospiral strains (Table 1) belonging to 7 pathogenic species and 5 non-pathogenic or saprophytic species were subjected to both PCR tests. All pathogenic leptospires were positive when tested with both assays. In addition, nested PCR amplified all leptospires including saprophytic and L. fainei and L. inadai species (two species whose pathogenicity is controversial). Conversely, our LFB1 assay only amplified the seven unambiguously pathogenic species. Additionally, fluorescence melting curve analysis showed specific discriminant melting temperatures for the leptospiral genomic species (Fig. 1). As an example, the mean melting temperature obtained for L. interrogans s.s. strain Verdun was 83.4 ± 0.7°C. To further determine the specificity of the assay, genomic DNA was purified and tested from cultures (laboratory isolates or collection strains) of a broad panel of organisms (Table 2). All of these non-leptospiral spp. were not amplified either by the LFB1 assay or by nested PCR. All PCR products of positive samples, visualized by gel electrophoresis (5 μl of each PCR product on a 1.5% agarose gel containing ethidium bromide), showed a unique 331-bp band. All negative samples showed little or no amplification caused by primer–dimer formation (Fig. 2).
Melting curve analysis of pathogenic Leptospira genomospecies after real-time amplification with LFB1 primers and SYBR Green dye in the LightCycler.
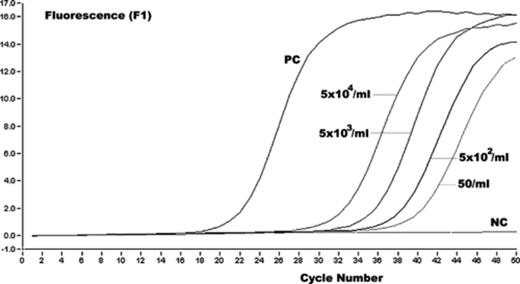
Representative results of Leptospira amplicon (strain Verdun) detection in channel F1. The fluorescence values versus cycle number are displayed. Serially diluted serum containing 5 × 104–50 leptospires/ml were used as standards. Two ng of purified DNA were used as positive control (PC). As a negative control (NC), the template DNA was replaced with PCR-grade water.
3.2 Sensitivity of the PCR assay
Rather than using tenfold dilutions of DNA in water, we preferred to test dilutions of a reference culture in a negative serum. By testing dilutions of a L. interrogans strain Verdun culture, ranging from 5 × 104 to 5 leptospires/ml of serum, the limit of sensitivity of the LFB1 assay was determined to be 50 leptospires/ml. The same sensitivity was obtained with the rrs nested-PCR. This minimum detection limit corresponds approximately to one to two genome copies in the PCR mixture. With both PCRs, a minimum detection limit of approximately 5 cells was established per reaction using artificially seeded urine samples.
3.3 Detection of leptospiral DNA in human clinical specimens
To assess the ability of our LFB1 real-time assay to detect the presence of pathogenic leptospires in humans, serum samples were collected from patients living in Pacific Island Countries and Territories (PICTs) with the Pacific Public Health Surveillance Network logistic in the Pacific Islands [[2]. From July 2004 to February 2005, 61 serum samples were obtained from 51 patients (mean age, 30 years) who were investigated for possible systemic leptospirosis. All patients (39 males, 12 females) originating from Marquesas islands (6 patients), Society islands (13 patients), Wallis and Futuna (18 patients) or New Caledonia (14 patients), presented with various symptoms compatible with this endemic disease (Table 3).
rrs nested PCR, LFB1 real-time assay and serologic results for 51 patients
| Patient no. | Origin | Sex | Age (years) | Days of samplinga | MAT titersb | Main serogroupc | rrs nested PCR | LFB1 real-time assay (leptospires/ml) | Species |
| 1 | Marquesas islands | M | 12 | 7 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 2 | Marquesas islands | M | 17 | 3 | 0 | − | − | ||
| 3 | Marquesas islands | M | 33 | 30 | 0 | − | − | ||
| 4 | Marquesas islands | M | 36 | 7 | 0 | − | − | ||
| 5 | Marquesas islands | M | 19 | 9 | 0 | − | − | ||
| 6 | Marquesas islands | M | 29 | 9 | 0 | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai | |
| 7 | Society islands | F | 38 | 5 | 0 | + | 2.5 × 102 | L. interrogans s.s.–L. santarosai | |
| 8 | Society islands | M | 8 | 2 | 0 | − | − | ||
| 9 | Society islands | M | 19 | 3 | 100 | Australis | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai |
| 10 | Society islands | M | 45 | 3 | 100 | Ictero. | − | − | |
| 11 | Society islands | F | 19 | 4 | 100 | Ictero. | − | − | |
| 12 | Society islands | F | 33 | 5 | 0 | − | − | ||
| 13 | Society islands | M | 57 | 4 | 0 | − | − | ||
| 14 | Society islands | M | 20 | 2 | 0 | + | 1.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 15 | Society islands | F | 68 | 8 | 0 | + | 5.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 16 | Society islands | M | 21 | 5 | 0 | + | 3.9 × 104 | L. interrogans s.s.–L. santarosai | |
| 17 | Society islands | M | 20 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | Society islands | M | 45 | 3 | 0 | + | 1.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | Society islands | M | 19 | 4 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 20 | Futuna | M | 18 | 1 | 0 | − | − | ||
| 21 | Futuna | F | 23 | 5 | 0 | − | − | ||
| 22 | Futuna | M | 15 | 13 | 100 | Ictero. | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai |
| 23d | Futuna | M | 15 | 6 | 100 | Australis | + | 1.2 × 102 | L. kirschneri–L. noguchii |
| 10 | 400 | Australis | − | − | |||||
| 24 | Futuna | M | 25 | 7 | 50 | Ictero. | − | − | |
| 25 | Futuna | F | 43 | 11 | 0 | − | − | ||
| 26 | Futuna | M | 14 | 12 | 100 | Autumnalis | − | − | |
| 27 | Futuna | M | 28 | 10 | 400 | Ictero. | − | − | |
| 28 | Futuna | M | 18 | 14 | 800 | Australis | − | − | |
| 29 | Futuna | M | 15 | NA | 0 | − | − | ||
| 30 | Futuna | M | 52 | 1 | 0 | − | − | ||
| 31 | Futuna | M | 44 | 1 | 0 | − | − | ||
| 32 | Futuna | F | 30 | 1 | 0 | − | − | ||
| 33 | Futuna | M | 31 | 1 | 0 | − | − | ||
| 34d | Futuna | F | 26 | 6 | 0 | + | 2.0 × 103 | L. kirschneri–L. noguchii | |
| 10 | 400 | Ictero. | − | − | |||||
| 35 | Wallis | F | 13 | 6 | 0 | − | − | ||
| 36 | Wallis | M | 47 | 8 | 0 | − | − | ||
| 37d | Wallis | M | 34 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 10 | 100 | Ictero. | − | − | |||||
| 38 | New Caledonia | M | 54 | 2 | 0 | + | 2.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 39d | New Caledonia | M | 38 | 5 | 0 | + | 3.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | 800 | Ictero. | − | − | |||||
| 40d | New Caledonia | M | 49 | 6 | 0 | + | 1.2 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | 200 | Autumnalis | − | − | |||||
| 41d | New Caledonia | M | 9 | 10 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 12 | 400 | Ictero. | − | − | |||||
| 42d | New Caledonia | M | 15 | 2 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 11 | 400 | Ictero. | − | − | |||||
| 43d | New Caledonia | M | 36 | 5 | 0 | + | 6.3 × 102 | L. interrogans s.s.–L. santarosai | |
| 28 | 400 | Ictero./Ballum | − | − | |||||
| 44 | New Caledonia | M | 37 | 10 | 0 | − | − | ||
| 45 | New Caledonia | F | 55 | 12 | 0 | − | − | ||
| 46 | New Caledonia | F | 33 | 5 | 0 | + | 6.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 47 | New Caledonia | M | 15 | 4 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 48 | New Caledonia | M | 35 | 1 | 0 | + | 2.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 49d | New Caledonia | M | 14 | 8 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 23 | 3200 | Ictero. | − | − | |||||
| 50 | New Caledonia | F | 56 | 14 | 0 | − | − | ||
| 51d | New Caledonia | M | 58 | 8 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 20 | 800 | Ictero. | − | − |
| Patient no. | Origin | Sex | Age (years) | Days of samplinga | MAT titersb | Main serogroupc | rrs nested PCR | LFB1 real-time assay (leptospires/ml) | Species |
| 1 | Marquesas islands | M | 12 | 7 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 2 | Marquesas islands | M | 17 | 3 | 0 | − | − | ||
| 3 | Marquesas islands | M | 33 | 30 | 0 | − | − | ||
| 4 | Marquesas islands | M | 36 | 7 | 0 | − | − | ||
| 5 | Marquesas islands | M | 19 | 9 | 0 | − | − | ||
| 6 | Marquesas islands | M | 29 | 9 | 0 | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai | |
| 7 | Society islands | F | 38 | 5 | 0 | + | 2.5 × 102 | L. interrogans s.s.–L. santarosai | |
| 8 | Society islands | M | 8 | 2 | 0 | − | − | ||
| 9 | Society islands | M | 19 | 3 | 100 | Australis | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai |
| 10 | Society islands | M | 45 | 3 | 100 | Ictero. | − | − | |
| 11 | Society islands | F | 19 | 4 | 100 | Ictero. | − | − | |
| 12 | Society islands | F | 33 | 5 | 0 | − | − | ||
| 13 | Society islands | M | 57 | 4 | 0 | − | − | ||
| 14 | Society islands | M | 20 | 2 | 0 | + | 1.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 15 | Society islands | F | 68 | 8 | 0 | + | 5.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 16 | Society islands | M | 21 | 5 | 0 | + | 3.9 × 104 | L. interrogans s.s.–L. santarosai | |
| 17 | Society islands | M | 20 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | Society islands | M | 45 | 3 | 0 | + | 1.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | Society islands | M | 19 | 4 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 20 | Futuna | M | 18 | 1 | 0 | − | − | ||
| 21 | Futuna | F | 23 | 5 | 0 | − | − | ||
| 22 | Futuna | M | 15 | 13 | 100 | Ictero. | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai |
| 23d | Futuna | M | 15 | 6 | 100 | Australis | + | 1.2 × 102 | L. kirschneri–L. noguchii |
| 10 | 400 | Australis | − | − | |||||
| 24 | Futuna | M | 25 | 7 | 50 | Ictero. | − | − | |
| 25 | Futuna | F | 43 | 11 | 0 | − | − | ||
| 26 | Futuna | M | 14 | 12 | 100 | Autumnalis | − | − | |
| 27 | Futuna | M | 28 | 10 | 400 | Ictero. | − | − | |
| 28 | Futuna | M | 18 | 14 | 800 | Australis | − | − | |
| 29 | Futuna | M | 15 | NA | 0 | − | − | ||
| 30 | Futuna | M | 52 | 1 | 0 | − | − | ||
| 31 | Futuna | M | 44 | 1 | 0 | − | − | ||
| 32 | Futuna | F | 30 | 1 | 0 | − | − | ||
| 33 | Futuna | M | 31 | 1 | 0 | − | − | ||
| 34d | Futuna | F | 26 | 6 | 0 | + | 2.0 × 103 | L. kirschneri–L. noguchii | |
| 10 | 400 | Ictero. | − | − | |||||
| 35 | Wallis | F | 13 | 6 | 0 | − | − | ||
| 36 | Wallis | M | 47 | 8 | 0 | − | − | ||
| 37d | Wallis | M | 34 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 10 | 100 | Ictero. | − | − | |||||
| 38 | New Caledonia | M | 54 | 2 | 0 | + | 2.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 39d | New Caledonia | M | 38 | 5 | 0 | + | 3.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | 800 | Ictero. | − | − | |||||
| 40d | New Caledonia | M | 49 | 6 | 0 | + | 1.2 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | 200 | Autumnalis | − | − | |||||
| 41d | New Caledonia | M | 9 | 10 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 12 | 400 | Ictero. | − | − | |||||
| 42d | New Caledonia | M | 15 | 2 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 11 | 400 | Ictero. | − | − | |||||
| 43d | New Caledonia | M | 36 | 5 | 0 | + | 6.3 × 102 | L. interrogans s.s.–L. santarosai | |
| 28 | 400 | Ictero./Ballum | − | − | |||||
| 44 | New Caledonia | M | 37 | 10 | 0 | − | − | ||
| 45 | New Caledonia | F | 55 | 12 | 0 | − | − | ||
| 46 | New Caledonia | F | 33 | 5 | 0 | + | 6.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 47 | New Caledonia | M | 15 | 4 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 48 | New Caledonia | M | 35 | 1 | 0 | + | 2.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 49d | New Caledonia | M | 14 | 8 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 23 | 3200 | Ictero. | − | − | |||||
| 50 | New Caledonia | F | 56 | 14 | 0 | − | − | ||
| 51d | New Caledonia | M | 58 | 8 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 20 | 800 | Ictero. | − | − |
aEstimated days after first clinical symptoms.
bAccording to usual interpretation, a titer≥100 was considered positive.
cIctero., icterohaemorrhagiae.
dSerologically confirmed cases were those with seroconversion or a titer rise≥2 dilutions between acute and convalescent phases.
rrs nested PCR, LFB1 real-time assay and serologic results for 51 patients
| Patient no. | Origin | Sex | Age (years) | Days of samplinga | MAT titersb | Main serogroupc | rrs nested PCR | LFB1 real-time assay (leptospires/ml) | Species |
| 1 | Marquesas islands | M | 12 | 7 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 2 | Marquesas islands | M | 17 | 3 | 0 | − | − | ||
| 3 | Marquesas islands | M | 33 | 30 | 0 | − | − | ||
| 4 | Marquesas islands | M | 36 | 7 | 0 | − | − | ||
| 5 | Marquesas islands | M | 19 | 9 | 0 | − | − | ||
| 6 | Marquesas islands | M | 29 | 9 | 0 | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai | |
| 7 | Society islands | F | 38 | 5 | 0 | + | 2.5 × 102 | L. interrogans s.s.–L. santarosai | |
| 8 | Society islands | M | 8 | 2 | 0 | − | − | ||
| 9 | Society islands | M | 19 | 3 | 100 | Australis | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai |
| 10 | Society islands | M | 45 | 3 | 100 | Ictero. | − | − | |
| 11 | Society islands | F | 19 | 4 | 100 | Ictero. | − | − | |
| 12 | Society islands | F | 33 | 5 | 0 | − | − | ||
| 13 | Society islands | M | 57 | 4 | 0 | − | − | ||
| 14 | Society islands | M | 20 | 2 | 0 | + | 1.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 15 | Society islands | F | 68 | 8 | 0 | + | 5.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 16 | Society islands | M | 21 | 5 | 0 | + | 3.9 × 104 | L. interrogans s.s.–L. santarosai | |
| 17 | Society islands | M | 20 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | Society islands | M | 45 | 3 | 0 | + | 1.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | Society islands | M | 19 | 4 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 20 | Futuna | M | 18 | 1 | 0 | − | − | ||
| 21 | Futuna | F | 23 | 5 | 0 | − | − | ||
| 22 | Futuna | M | 15 | 13 | 100 | Ictero. | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai |
| 23d | Futuna | M | 15 | 6 | 100 | Australis | + | 1.2 × 102 | L. kirschneri–L. noguchii |
| 10 | 400 | Australis | − | − | |||||
| 24 | Futuna | M | 25 | 7 | 50 | Ictero. | − | − | |
| 25 | Futuna | F | 43 | 11 | 0 | − | − | ||
| 26 | Futuna | M | 14 | 12 | 100 | Autumnalis | − | − | |
| 27 | Futuna | M | 28 | 10 | 400 | Ictero. | − | − | |
| 28 | Futuna | M | 18 | 14 | 800 | Australis | − | − | |
| 29 | Futuna | M | 15 | NA | 0 | − | − | ||
| 30 | Futuna | M | 52 | 1 | 0 | − | − | ||
| 31 | Futuna | M | 44 | 1 | 0 | − | − | ||
| 32 | Futuna | F | 30 | 1 | 0 | − | − | ||
| 33 | Futuna | M | 31 | 1 | 0 | − | − | ||
| 34d | Futuna | F | 26 | 6 | 0 | + | 2.0 × 103 | L. kirschneri–L. noguchii | |
| 10 | 400 | Ictero. | − | − | |||||
| 35 | Wallis | F | 13 | 6 | 0 | − | − | ||
| 36 | Wallis | M | 47 | 8 | 0 | − | − | ||
| 37d | Wallis | M | 34 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 10 | 100 | Ictero. | − | − | |||||
| 38 | New Caledonia | M | 54 | 2 | 0 | + | 2.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 39d | New Caledonia | M | 38 | 5 | 0 | + | 3.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | 800 | Ictero. | − | − | |||||
| 40d | New Caledonia | M | 49 | 6 | 0 | + | 1.2 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | 200 | Autumnalis | − | − | |||||
| 41d | New Caledonia | M | 9 | 10 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 12 | 400 | Ictero. | − | − | |||||
| 42d | New Caledonia | M | 15 | 2 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 11 | 400 | Ictero. | − | − | |||||
| 43d | New Caledonia | M | 36 | 5 | 0 | + | 6.3 × 102 | L. interrogans s.s.–L. santarosai | |
| 28 | 400 | Ictero./Ballum | − | − | |||||
| 44 | New Caledonia | M | 37 | 10 | 0 | − | − | ||
| 45 | New Caledonia | F | 55 | 12 | 0 | − | − | ||
| 46 | New Caledonia | F | 33 | 5 | 0 | + | 6.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 47 | New Caledonia | M | 15 | 4 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 48 | New Caledonia | M | 35 | 1 | 0 | + | 2.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 49d | New Caledonia | M | 14 | 8 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 23 | 3200 | Ictero. | − | − | |||||
| 50 | New Caledonia | F | 56 | 14 | 0 | − | − | ||
| 51d | New Caledonia | M | 58 | 8 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 20 | 800 | Ictero. | − | − |
| Patient no. | Origin | Sex | Age (years) | Days of samplinga | MAT titersb | Main serogroupc | rrs nested PCR | LFB1 real-time assay (leptospires/ml) | Species |
| 1 | Marquesas islands | M | 12 | 7 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 2 | Marquesas islands | M | 17 | 3 | 0 | − | − | ||
| 3 | Marquesas islands | M | 33 | 30 | 0 | − | − | ||
| 4 | Marquesas islands | M | 36 | 7 | 0 | − | − | ||
| 5 | Marquesas islands | M | 19 | 9 | 0 | − | − | ||
| 6 | Marquesas islands | M | 29 | 9 | 0 | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai | |
| 7 | Society islands | F | 38 | 5 | 0 | + | 2.5 × 102 | L. interrogans s.s.–L. santarosai | |
| 8 | Society islands | M | 8 | 2 | 0 | − | − | ||
| 9 | Society islands | M | 19 | 3 | 100 | Australis | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai |
| 10 | Society islands | M | 45 | 3 | 100 | Ictero. | − | − | |
| 11 | Society islands | F | 19 | 4 | 100 | Ictero. | − | − | |
| 12 | Society islands | F | 33 | 5 | 0 | − | − | ||
| 13 | Society islands | M | 57 | 4 | 0 | − | − | ||
| 14 | Society islands | M | 20 | 2 | 0 | + | 1.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 15 | Society islands | F | 68 | 8 | 0 | + | 5.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 16 | Society islands | M | 21 | 5 | 0 | + | 3.9 × 104 | L. interrogans s.s.–L. santarosai | |
| 17 | Society islands | M | 20 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | Society islands | M | 45 | 3 | 0 | + | 1.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | Society islands | M | 19 | 4 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 20 | Futuna | M | 18 | 1 | 0 | − | − | ||
| 21 | Futuna | F | 23 | 5 | 0 | − | − | ||
| 22 | Futuna | M | 15 | 13 | 100 | Ictero. | + | 8.0 × 101 | L. interrogans s.s.–L. santarosai |
| 23d | Futuna | M | 15 | 6 | 100 | Australis | + | 1.2 × 102 | L. kirschneri–L. noguchii |
| 10 | 400 | Australis | − | − | |||||
| 24 | Futuna | M | 25 | 7 | 50 | Ictero. | − | − | |
| 25 | Futuna | F | 43 | 11 | 0 | − | − | ||
| 26 | Futuna | M | 14 | 12 | 100 | Autumnalis | − | − | |
| 27 | Futuna | M | 28 | 10 | 400 | Ictero. | − | − | |
| 28 | Futuna | M | 18 | 14 | 800 | Australis | − | − | |
| 29 | Futuna | M | 15 | NA | 0 | − | − | ||
| 30 | Futuna | M | 52 | 1 | 0 | − | − | ||
| 31 | Futuna | M | 44 | 1 | 0 | − | − | ||
| 32 | Futuna | F | 30 | 1 | 0 | − | − | ||
| 33 | Futuna | M | 31 | 1 | 0 | − | − | ||
| 34d | Futuna | F | 26 | 6 | 0 | + | 2.0 × 103 | L. kirschneri–L. noguchii | |
| 10 | 400 | Ictero. | − | − | |||||
| 35 | Wallis | F | 13 | 6 | 0 | − | − | ||
| 36 | Wallis | M | 47 | 8 | 0 | − | − | ||
| 37d | Wallis | M | 34 | 3 | 0 | + | 7.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 10 | 100 | Ictero. | − | − | |||||
| 38 | New Caledonia | M | 54 | 2 | 0 | + | 2.0 × 103 | L. interrogans s.s.–L. santarosai | |
| 39d | New Caledonia | M | 38 | 5 | 0 | + | 3.9 × 102 | L. interrogans s.s.–L. santarosai | |
| 18 | 800 | Ictero. | − | − | |||||
| 40d | New Caledonia | M | 49 | 6 | 0 | + | 1.2 × 103 | L. interrogans s.s.–L. santarosai | |
| 19 | 200 | Autumnalis | − | − | |||||
| 41d | New Caledonia | M | 9 | 10 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 12 | 400 | Ictero. | − | − | |||||
| 42d | New Caledonia | M | 15 | 2 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 11 | 400 | Ictero. | − | − | |||||
| 43d | New Caledonia | M | 36 | 5 | 0 | + | 6.3 × 102 | L. interrogans s.s.–L. santarosai | |
| 28 | 400 | Ictero./Ballum | − | − | |||||
| 44 | New Caledonia | M | 37 | 10 | 0 | − | − | ||
| 45 | New Caledonia | F | 55 | 12 | 0 | − | − | ||
| 46 | New Caledonia | F | 33 | 5 | 0 | + | 6.3 × 103 | L. interrogans s.s.–L. santarosai | |
| 47 | New Caledonia | M | 15 | 4 | 0 | + | 7.9 × 103 | L. interrogans s.s.–L. santarosai | |
| 48 | New Caledonia | M | 35 | 1 | 0 | + | 2.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 49d | New Caledonia | M | 14 | 8 | 0 | + | 2.5 × 103 | L. interrogans s.s.–L. santarosai | |
| 23 | 3200 | Ictero. | − | − | |||||
| 50 | New Caledonia | F | 56 | 14 | 0 | − | − | ||
| 51d | New Caledonia | M | 58 | 8 | 0 | + | 5.0 × 102 | L. interrogans s.s.–L. santarosai | |
| 20 | 800 | Ictero. | − | − |
aEstimated days after first clinical symptoms.
bAccording to usual interpretation, a titer≥100 was considered positive.
cIctero., icterohaemorrhagiae.
dSerologically confirmed cases were those with seroconversion or a titer rise≥2 dilutions between acute and convalescent phases.
The biological course of the disease was followed for 1–30 days (average, 8 days) after the estimated onset of illness (day 0). Both PCR assays confirmed the diagnosis for 25 patients (49%, 25/51). Furthermore, our LFB1 real-time assay revealed an estimated density of 8.0 × 0101–3.9 × 104 leptospires per ml of blood. The average density (1 × 103/ml) was lower than the previously estimated critical threshold of 1 × 104/ml [[]. Only one patient (16) exhibited a “high” density of leptospires (3.9 × 104/ml) and clinically the vital prognosis was good.
Seroconversion was observed in 9 patients (34, 37, 39 to 43, 49 and 51) at a median of 17 days after the appearance of symptoms. The PCR signal was invariably positive for the first serum sample (which was negative by serology), giving unequivocal confirmation of leptospirosis. This finding is corroborated by previous reports of early detection of leptospires in blood [[3] and a progressive decrease of leptospiral density due to host antibody response. Increasing leptospiral antibody titers were observed for one patient (23). A positive PCR signal was obtained for the first serum sample and became negative as the antibody titers rose.
Single acute samples were collected from 41 patients showing negative MAT titers (34 patients) or low antibody titers (7 patients). According to usual interpretation, the positive threshold of the MAT reaction is fixed at 1/100 and titers between 1/100 and 1/400 are considered positive but non significative. The presence of leptospiral DNA was demonstrated in the sera of 11 patients, mainly those with negative MAT titers (82%, 9/11).
Out of 25 positive samples, 23 exhibited a melting curve compatible with L. interrogans s.s. or L. santarosai and 2 with L. kirschneri or L. noguchii. The total assay time for 12 clinical specimens, including DNA extraction and PCR amplification, was less than 3 hours, after which the data could be analysed. In comparison, 24 hours were necessary to complete a rrs nested-PCR.
These data illustrate the potential of our LFB1 real-time assay for the detection of leptospires in serum samples and their subsequent quantification in a single run. The practicability of the method makes it suitable for use in prospective studies of human leptospirosis cases.
4 Discussion
In diagnostic laboratories, detection of pathogenic leptospires by real-time PCR offers considerable benefits compared to conventional methods. This technology enables PCR to be performed with greatly reduced carry-over contamination risk and with minimal hands-on time. Use of SYBR Green I dye provides a simple and inexpensive real-time PCR detection technique. Although melting curve analysis with SYBR Green is sometimes considered less specific compared to the use of fluorescent probes [[4], the need of more expensive probes is not always necessary when conditions of amplification have been correctly optimized (especially primer sequences and concentrations). The specific product melting peaks with no primer–dimer or other non-specific product signal provided evidence that our assay is specific. By optimizing the different steps to be performed we also decreased the cost of performing LFB1 test which is in the same range of that of traditional PCR assays. Our turnaround time was considerably faster with the new real-time PCR assay. Occasionally, serum samples arriving late in the morning were treated in emergency and results were transmitted to the physician the same day. This compares favourably to the extra day necessary with the nested-PCR. Our in-house LFB1 PCR assay without any post-amplification procedures could be adapted to other real-time PCR test systems such as the Roche TaqMan 48 machine.
The possibility to quantitate leptospire density accurately in patients will provide an objective evaluation of antibiotics efficiency. Although beta-lactams are first choice antibiotics for treatment of acute leptospirosis, persistence of leptospires in humans after the initial clinical period [[3,[5] suggests that they are ineffective in clearing leptospires that are located in protected sites [[6]. The use of other antibiotic regimens (deoxycycline, ofloxacin) could be required to definitively eradicate leptospirosis from the host [[].
Our real-time PCR only amplified the 7 unambiguously pathogenic Leptospira species but did not detect non-pathogenic strains or other pathogens (Table 2). The real-time PCR system proposed by Levett et al. [[7] has the same specificity as our LFB1 assay. This is a major improvement as Levett et al. demonstrated a limitation in our previous PCR assay [[]. Indeed, primers described for the amplification of the rrs gene [[] detected both pathogenic and saprophytic strains (L. biflexa), which in the unlikely event of contamination of specimens with environmental strains might produce a false-positive result. However, in our experience, in practice this potential shortcoming was very limited (if at all) in clinical material. Smythe et al. [[4] developed a quantitative PCR (TaqMan) assay for pathogenic leptospires but amplification products were obtained for L. meyeri (serovar perameles strain bandicoot 343) and one strain of L. inadai.
A limitation of PCR-based diagnosis of leptospirosis is the inability of most assays to identify the infecting strain at the species, serogroup or serovar level. Indeed, leptospiral species can be identified either by electrophoresis in nondenaturing polyacrylamide gels [[8] or amplicon sequencing [[]. However, these methods are time consuming. The ability of the LightCycler PCR to distinguish between pathogenic species was possible using the melting curves, thus providing a fast alternative for identification. Indeed, within a range of 3°C, it was possible to assign a specific Tm to a single species or a set of species. L. alexanderi and L. borgpetersenii showed the mean highest Tm (86.4°C) while the mean lowest Tm (83.4°C) was observed with L. interrogans s.s. and L. santarosai. A single specific Tm (85.2°C) was observed with L. weilii. Although the LipL32 Leptospira real-time PCR proposed by Levett et al. [[7] was able to delineate such pathogenic species, the mean melting curve temperature for the leptospires tested ranged from 83.6 to 85°C (range 1.4°C), giving a slightly lower resolution assay. Indeed, L. interrogans, L. kirschneri and L. noguchii had the same Tm (84.0°C) while our LFB1 real-time assay clearly discriminated L. interrogans (83.4°C) from L. kirschneri and L. noguchii (84.4°C). In any case, distinction between L. interrogans and L. noguchii on one hand and L. borgpetersenii and L. santarosai on the other is not an actual problem. Indeed, although L. interrogans, L. kirschneri and L. borgpetersenii are ubiquitous species [[,[], L. santarosai and L. noguchii are mainly found in South America while L. alexanderi and L. weilii are isolated in Asia. In New Caledonia, Leptospira strains isolated from human patients and identified at serovar level belonged to L. interrogans, L. kirschneri or L. borgpetersenii. Similarly, in the past few years rrs fragment amplicons from 25 patients have been sequenced to determine the infecting species and 22 sequences were assigned to L. interrogans, 1 to L. kirschneri and 3 to L. borgpetersenii (unpublished data). Therefore, we can assume that species identified among the 19 positive patients evidenced using our real-time assay were L. interrogans and L. kirschneri, which are representative of the main circulating species in New Caledonia. Similar conclusions would be deduced in continental France, and probably in Europe, where only L. interrogans, L. kirschneri and L. borgpetersenii are encountered in human patients (unpublished data).
In conclusion, we have developed a real-time PCR assay based on SYBR Green technology for highly specific detection of pathogenic leptospires in clinical samples. This assay delineates pathogenic genomospecies and can detect as few as 50 leptospires/ml of serum. This method is simple, rapid, and has applications in human and veterinary fields as both research and diagnostic testing.
Acknowledgements
Thanks are due to J.F. Yvon and C. Coudert for the samples from Wallis and Futuna and French Polynesia. The authors are indebted to J.F. Mackay for editorial contributions. This work was supported by grant from Pasteur-CERBA.
References