Abstract
Background:
To develop and validate a nomogram and web-based calculator to predict overall survival (OS) in Caucasian-advanced oesophagogastric adenocarcinoma (AOA) patients undergoing first-line combination chemotherapy.
Methods:
Nine hundred twenty-four AOA patients treated at 28 Spanish teaching hospitals from January 2008 to September 2014 were used as derivation cohort. The result of an adjusted-Cox proportional hazards regression was represented as a nomogram and web-based calculator. The model was validated in 502 prospectively recruited patients treated between October 2014 and December 2016. Harrell's c-index was used to evaluate discrimination.
Results:
The nomogram includes seven predictors associated with OS: HER2-positive tumours treated with trastuzumab, Eastern Cooperative Oncology Group performance status, number of metastatic sites, bone metastases, ascites, histological grade, and neutrophil-to-lymphocyte ratio. Median OS was 5.8 (95% confidence interval (CI), 4.5–6.6), 9.4 (95% CI, 8.5–10.6), and 14 months (95% CI, 11.8–16) for high-, intermediate-, and low-risk groups, respectively (P<0.001), in the derivation set and 4.6 (95% CI, 3.3–8.1), 12.7 (95% CI, 11.3–14.3), and 18.3 months (95% CI, 14.6–24.2) for high-, intermediate-, and low-risk groups, respectively (P<0.001), in the validation set. The nomogram is well-calibrated and reveals acceptable discriminatory capacity, with optimism-corrected c-indices of 0.618 (95% CI, 0.591–0.631) and 0.673 (95% CI, 0.636–0.709) in derivation and validation groups, respectively. The AGAMENON nomogram outperformed the Royal Marsden Hospital (c-index=0.583; P=0.00046) and Japan Clinical Oncology Group prognostic indices (c-index=0.611; P=0.03351).
Conclusions:
We developed and validated a straightforward model to predict survival in Caucasian AOA patients initiating first-line polychemotherapy. This model can contribute to inform clinical decision-making and optimise clinical trial design.
Similar content being viewed by others
Main
Although the incidence of advanced oesofagogastric adenocarcinoma (AOA) has declined over the recent few years, it remains the fifth most common malignancy worldwide and the third cause of cancer-related death in both sexes (Ferlay et al, 2015). In general, combination chemotherapy (CT) regimens based on a platinum and fluoropyrimidine doublet with or without a third drug are the mainstay of treatment, yielding response rates of 40–45%. Nonetheless, they translate into only modest progression-free survival (PFS) and overall survival (OS), which rarely exceed 7 and 12 months, respectively, in HER2-negative tumours (Wagner et al, 2006). Tumours overexpressing or amplifying HER2 constitute a distinct subgroup, as adding trastuzumab to cisplatin and fluoropyrimidines has shown a clear survival benefit (Bang et al, 2010).
Other molecular targeted therapies have also been investigated, mostly with disappointing results (Ohtsu et al, 2011; Waddell et al, 2013), highlighting the need for reliable biomarkers to predict treatment effectiveness. Similarly, AOA’s unfavourable prognosis and the current interest in precision medicine emphasise the imperative for prognostic tools that can identify patients who would derive little survival benefit from modern therapeutic strategies and should therefore be actively recruited into clinical trials evaluating alternative treatments. In recent years, two prognostic indices to predict OS in patients with AOA receiving first-line CT based on clinical and laboratory attributes have been prospectively validated: the Royal Marsden Hospital prognostic index (RMH-I; Chau et al, 2004) and the Japan Clinical Oncology Group prognostic index (JCOG-I; Takahari et al, 2014). However, both were developed prior to HER2 testing and front-line trastuzumab for HER2-positive tumours became standard. Moreover, validations were carried out in the context of randomised clinical trials, far from ideal for prognostic factor analysis, as samples are conditioned by inclusion criteria and may not be representative of the overall AOA population, with patients who are usually older, suffer chronic comorbidities, or have a worse performance status.
With this background, we sought to develop and validate a nomogram, and online calculator to predict OS for Caucasian patients with advanced distal oesophagus, gastroesophageal junction (GEJ), or gastric adenocarcinoma undergoing first-line combination CT in the era of trastuzumab.
Patients and methods
Study design and population
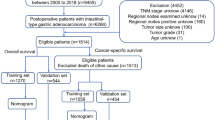
AGAMENON is a multicenter study that compiles consecutive cases of AOA patients from 28 Spanish teaching hospitals. Descriptions of the design, data collection, quality and eligibility criteria have been previously reported (Carmona-Bayonas et al, 2016; Jiménez-Fonseca et al, 2017). Briefly, patients eligible for inclusion were adults (age ⩾18 years) diagnosed with pathologically confirmed unresectable locally advanced or metastatic adenocarcinoma of the distal oesophagus, GEJ, and stomach who received at least one cycle of polychemotherapy using two or more drugs as first-line treatment. Patients for whom the information needed to complete the database could not be obtained, patients participating in a clinical trial in which no standard CT was used, those for whom follow-up was <3 months (except individuals with early demise during this period who had already been included), <6 months since completion of prior neoadjuvant or adjuvant treatment, and other primary and synchronous metastatic malignancy were excluded. The nomogram was derived in a cohort of consecutive AOA patients treated between January 2008 and September 2014 (Custodio et al, 2016). Here, we have validated our model in an independent cohort with similar eligibility criteria prospectively recruited from October 2014 to December 2016.
Variables
To elaborate the prognostic nomogram, we considered 29 routinely available baseline clinicopathological variables proven to predict survival in at least one previous publication. The analyses included: (1) patient-related variables such as age, gender, Eastern Cooperative Oncology Group Performance Status (ECOG-PS) scale, number of chronic comorbidities present prior to the first cycle of CT according to the Charlson comorbidity index (Charlson et al, 1994); haematological parameters including haemoglobin, white cell count, neutrophil-to-lymphocyte ratio (NLR), and platelet count; biochemical analytes including bilirubin, albumin, alkaline phosphatase, lactate dehydrogenase, and carcinoembryonic antigen (CEA); (2) tumour-dependent characteristics, such as stage at first diagnosis, primary tumour site, number of metastatic sites (organs involved), specific metastases (liver, lung, distant lymph nodes, peritoneum, ascites according to the European Association for the Study of the Liver (EASL) clinical practice guidelines (European Association for the Study of the Liver, 2010), bone), surgery of primary tumour, peri-operative treatment in tumours diagnosed in non-metastatic stages that subsequently recurred and the presence of serious, acute, cancer-derived complications at advanced disease diagnosis, such as intestinal obstruction, massive ascites, major bleeding, thromboembolic disease, and liver, respiratory, or renal dysfunction; (3) histological features such as grade, Lauren classification, presence of signet-ring cells, and HER2 status. To represent HER2-positive subtypes, we opted pragmatically to use the predictor ‘HER2-positive tumours treated with trastuzumab’, because in modern series of metastatic gastric cancer, the effect of HER2 overexpression or amplification is expected to be coupled to the use of anti-HER2 therapy. Laboratory variables were dichotomised with the cutoff at the limit of its normal range at each institution and had to be acquired within the 10 days preceding the first cycle of CT according to the local protocols of the participating centres. Primary tumour site was coded as oesophageal for Siewert I and II distal oesophageal tumours, whereas the remaining tumour locations were coded as gastric neoplasms. For the total number of metastatic sites, each lymph node region was considered independently. Unjustified missing values were not allowed, except for histopathological variables not determined in the course of ordinary care.
The primary endpoint was OS, which was estimated using the Kaplan–Meier method and defined as the time elapsed from the date of the first cycle of front-line CT to the date of death from any cause.
The information was gathered and updated from medical records or directly from the patient by medical oncologists experienced in treating AOA and trained to comply with the study requirements through a web-based platform (www.agamenonstudy.com). This tool includes real-time alerts to prevent inconsistencies, unjustified missing values, and errors. An independent researcher (MLSL) also carried out telephone monitoring.
The study was approved by a multicenter Research Ethics Committee. All patients still alive at the time of data collection provided written signed informed consent.
Statistical considerations
The development of the prognostic model started with a univariate assessment of the effect of each clinicopathological parameter using a Cox proportional hazards (PH) regression model. Variables univariately predictive of OS (P<0.1) were entered into the multivariate Cox PH regression. We dropped the presence of signet-ring cells and peritoneum metastases after correlation analyses to avoid multicollinearity with Lauren classification and ascites, respectively. When histopathological variables presented missing values, we applied multiple imputation using predictive mean matching, as suggested in the literature (Harrell, 2015). A final Cox PH model was selected using a backward step-down procedure based on the Akaike information criterion (AIC) as a stopping rule. The PH assumption for each predictor was verified using the Schoenfeld residuals test (see definition in Supplementary Table 1).
A nomogram is a graphic representation of the solution of an equation that provides a reasonable approximation of the probability of a particular outcome. On the basis of the coefficients of the multivariable analysis, a nomogram to provide visualised estimation of individual OS probability at 1 and 2 years was formulated by R software. The same was carried out to develop an online calculator: http://www.iricom.es/prognostictools/agamenon/inicio.aspx.
To determine discrimination ability, we developed 1000 bootstrap replications that were used as internal validation subsets to estimate the bias-corrected concordance index (c-index). Calibration at 1-year was analysed by plotting observed vs predicted OS probabilities at 1-year and with the Gronnesby–Borgan goodnest-of-fit test (reported as the score test P-value). We stratified patients further into three risk groups, according to prespecified cutoff values of the predicted probability of 1-year OS (<25%, 25–50%, >50%), and plotted the respective Kaplan–Meier survival curves. The log-rank test was used to analyse survival functions. These procedures were utilised in both the training and validation sets. Based on previous research, a sample size of 502 patients with >100 events per variable would be sufficient for this validation analysis (Vergouwe et al, 2005). We also contrasted the discriminatory ability of our model against the RMH-I and the JCOG-I scales by examining c-indices in the validation cohort (Kang et al, 2015). Finally, a sensitivity analysis according to HER2 status was carried out using the entire dataset.
All statistical assessments were two-sided and P-values <0.05 were deemed statistically significant. Statistical analyses were performed using Stata 14 (Stata, College Station, TX, USA) and R software, version 3.3.1 (http://www.r-project.org), including the rms and compareC packages (Harrell et al, 2015; Kang et al, 2015).
Results
Baseline characteristics
At the time of analysis (March 2017), 2147 patients were screened, 1426 of whom met eligibility criteria (924 and 502 in the training and validation sets, respectively). Figure 1 outlines the recruitment process. The main descriptive statistics are presented in Table 1. There are virtually no differences in baseline characteristics between the derivation and validation sets. The most notable contrast is evidenced in the change in treatment patterns over time, with more HER2 testing and trastuzumab treatment, and less CT triplets in the validation series.
Outcomes
A total of 856 (92.6%) and 298 (59.3%) events were recorded in the training and validation cohorts, respectively. Median OS in the two groups was 9.8 (95% confidence interval (CI), 9.2–10.4) and 12.7 months (95% CI, 11.3–14), respectively. One- and two-year survival was 40.4% (95% CI, 37.3–43.5%) and 15.7% (95% CI, 13.3–20.4%) in the derivation set and 52.9% (95% CI, 48.3–57.9%) and 25.4% (95% CI, 20.6–31.2%) in the validation cohort.
Development of the nomogram
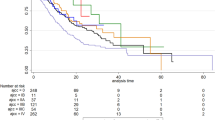
After the univariate analyses (Supplementary Table 2), multivariate survival analysis using Cox’s regression model was fitted on the training subset. Seven covariates correlated significantly with survival: Eastern Cooperative Oncology Group Performance Status (ECOG-PS), histological grade, number of metastatic sites, bone metastases, ascites, NLR, and HER2-positive tumours treated with trastuzumab (Table 2). We then devised a web-based calculator and prognostic nomogram to estimate the probability of 1- and 2-year OS (Figure 2). Schoenfeld’s global test, applied to verify the proportional hazards assumption, is presented in Supplementary Figure 1. The Gronnesby–Borgan score test demonstrated the model’s good fit in the derivation subset (χ2=6.639, P=0.15). The calibration plot displayed excellent agreement between the observed and expected probabilities of 1-year OS (Figure 3A). Across the 1000 bootstrap replications, the optimism-corrected c-index was 0.618 (95% CI, 0.591–0.631). Survival curves across prespecified cutoff values of the predicted probability of 1-year OS are shown in Figure 4A. Median OS was 5.8 months (95% CI, 4.5–6.6), 9.4 months (95% CI, 8.5–10.6), and 14 months (95% CI, 11.8–16) for the high-, intermediate-, and low-risk groups, respectively (log-rank P<0.001).
Kaplan-Meier survival curves. Overall survival curves according to the AGAMENON nomogram in the training subset (N=924) (A) and in the validation subset (complete case analysis, N=412) (B). In the validation subset the effective sample size was N=412 because complete case analysis was applied. Survival curves were stratified by the predicted survival probability at 12 months. Log-rank test, P<0.0001.
Validation of the nomogram
The nomogram’s performance was then assessed in the prospective cohort (complete cases analysis, n=412). A c-index of 0.673 (95% CI, 0.636–0.709) was observed. The nomogram prediction for 1-year OS also appeared to be well-calibrated in this subset (Gronnesby–Borgan score test: χ2=3.837, P=0.428; Figure 3B). Kaplan–Meier curves stratified on the basis of 1-year survival predictions are presented in Figure 4B. Median OS was 4.6 months (95% CI, 3.3–8.1), 12.7 months (95% CI, 11.3–14.3), and 18.3 months (95% CI, 14.6–24.2), for the high-, intermediate-, and low-risk groups, respectively (log-rank P<0.001).
Sensitivity analysis regarding the HER2 status
Sensitivity analyses showed that the ability of our nomogram to discriminate among prognostic categories in the entire dataset was seemingly homogeneous for HER2-negative and positive tumours (c-index of 0.623 (95% CI, 0.601–0.645) and 0.675 (95% CI, 0.629–0.722), respectively). Moreover, a simplified model with six predictors (excluding HER2-positive tumours treated with trastuzumab) was also well-calibrated in both HER2-negative and positive strata (calibration graphs by HER2 status not shown).
Comparison of AGAMENON nomogram, RMH, and JCOG
We also gauged the performance of the RMH-I and JCOG scores to our validation dataset. Both prognostic models were applied to those patients with available data for the seven parameters included in our nomogram (n=412). Although they are less accurate than the AGAMENON nomogram, both are able to discriminate survival outcomes for the three proposed categories in the validation subset (Supplementary Figures 2 and 3). The c-indices were 0.583 (95% CI, 0.545–0.621) for RMH-I and 0.611 (95%, CI 0.573–0.648) for JCOG-I, with P=0.00046 when AGAMENON and RMH-I were compared, P=0.03351 for AGAMENON vs JCOG-I, and P=0.17881 for RMH-I against JCOG-I.
Discussion
Several previous prognostic models based on clinicopathological variables have predicted survival in AOA patients undergoing palliative CT (Lee et al, 2007; Kim et al, 2008; Chau et al, 2009; Hsieh et al, 2016; Kawakami et al, 2016; Takahari et al, 2017; Wang et al, 2016). However, to the best of our knowledge, this is the first report of a nomogram developed and prospectively validated in a large series of Caucasian patients treated in the era of trastuzumab. We have demonstrated the AGAMENON nomogram’s ability to predict 1- and 2-year OS probability and its superior performance vs the RMH-I (Chau et al, 2004) and the JCOG-I models (Takahari et al, 2014).
AGAMENON uses readily available clinicopathological and laboratory covariates. Of them, poor PS (Chau et al, 2004; Lee et al, 2007; Kim et al, 2008; Takahari et al, 2014; Kawakami et al, 2016), the number of metastatic sites (Kim et al, 2008; Takahari et al, 2014; Wang et al, 2016), bone metastases (Lee et al, 2007; Kim et al, 2008) and peritoneal metastases (Chau et al, 2004; Kim et al, 2008), or malignant ascites (Lee et al, 2007) as a surrogate for peritoneal disease, are prognostic factors in common with some of the previously reported prognostic tools. High NLR status, a well-known biomarker of cancer-associated inflammation, has also exhibited a significant negative prognostic effect in many advanced malignancies, including large cohorts of gastric cancer patients (Grenader et al, 2016; Ock et al, 2016), and has been included in two recently published prognostic models in patients with advanced disease (Hsieh et al, 2016; Wang et al, 2016). However, the reader should be mindful that the effect of the NLR should be interpreted with caution in patients with intercurrent infectious processes, or in those undergoing corticosteroid therapy. On the other hand, although increasing American Joint Committee on Cancer (AJCC) histologic grade has been associated with decreasing survival in some reports (Sun et al, 2014; Duma et al, 2016), it has not emerged as an independent prognostic factor in any of the prognostic scales that have contemplated it in their analyses. As for HER2 status, most previous prognostic models were developed before HER2 testing and trastuzumab therapy became standard and two most recent studies conducted in the era of trastuzumab have not analysed its prognostic role (Hsieh et al, 2016; Wang et al, 2016). A retrospective study published in abstract form only (Kawakami et al, 2016) has derived and retrospectively validated a prognostic nomogram in Japanese metastatic gastric cancer patients starting first-line treatment that includes the HER2 status as predictor regardless of trastuzumab therapy. To our knowledge, AGAMENON is the only nomogram able to appraise the additive effect of HER2-positive tumours treated with trastuzumab together with other predictors in Caucasian AOA patients undergoing first-line CT, confirming that it increases survival in this population. Moreover, the model proved to be robust for both HER2-positive and negative tumours.
The two most commonly used prognostic indices developed on the foundation of large databases are the RMH-I and JCOG-I for Caucasian and Asian populations, respectively (Chau et al, 2004; Takahari et al, 2014). Both models were derived and externally validated using data from patients enroled in multicenter phase III clinical trials, which selection criteria tend to be restrictive, resulting in outcomes that may be not easily extrapolated to all patient groups. When we applied RMH-I and JCOG-I to our validation cohort, both models’ predictive performance was worse than that of the AGAMENON nomogram, probably because they fail to consider the effect of key variables in AOC (e.g., the HER2 status) and because their discriminatory ability is a function of the populations in which they were developed. Despite this, median OS for each risk category after applying the AGAMENON nomogram, RMH-I, or JCOG-I was better in our dataset than OS reported in the original studies.
The AGAMENON nomogram has several strengths. It was constructed within the framework of a large cohort of Caucasian AOA patients with a wide variety of variables available at diagnosis. The seven parameters included in our scale are confirmed prognostic factors that reflect the general status of the patient and disease, thereby providing clinically relevant information and accordingly enhancing the relevance of the tool developed. The model is statistically robust and respects pertinent quality criteria proposed for prognostic scores, such as a well-defined cohort of patients, a comprehensive evaluation of all well-established prognostic variables, reliable outcome measures, few patients lost to follow-up, and data available for the vast majority (Maltoni et al, 2005). Some imbalances are seen regarding clinical features, CT schedules, HER2 testing, and trastuzumab therapy, as well as survival between the derivation and validation subsets as a reflection of recent epidemiological trends and the changes in patient interventions over time (Jiménez-Fonseca et al, 2017). Moreover, as it usually happens in other similar registries, there were some biochemical variables in the prospective series with a small proportion of missing data: alkaline phosphatase (3.58%), lactate dehydrogenase (14.74%), bilirubin (0.60%), albumin (8.76%) and carcinoembryonic antigen (13.34%). However, it should be emphasised about this point that our nomogram’s ability to predict survival in the two series, despite these differences, reinforces the robustness of our findings and their applicability to other populations. A key aspect to consider is the different length of follow-up in the derivation series compared to the prospective validation cohort, which continued the patient recruitment at the time of analysis. Nevertheless, it does not seemingly affect the assessment of the discriminative capacity of the nomogram, insofar as the number of events necessary to validate this prognostic model has already been reached in the prospective series. Similarly, it also has no impact on the nomogram predictions, which are derived exclusively from mature data from the derivation cohort.
Several points should be taken into account when considering the generalisability of the model. First, although explanatory covariables appear to be appropriate in different contexts, one must always be mindful of the possibility of uncommon factors which could significantly affect the prognosis in a minority of patients, as well as those that might eventually develop during the course of the disease, as only baseline variables have been contemplated. In addition, an important point to clarify is that the prognostic categories of this nomogram were not used in the study to influence medical decision-making. Second, prognostic variables have been categorised or dichotomised according to previously published criteria and after proving a significant effect in the univariate analysis. However, it is obvious that it could be detrimental to the accuracy of outcome prediction. Third, although using the prognostic model is superior to decisions based on clinical experience and significantly improves the results of previous studies, we must bear in mind that its discriminatory capacity is restricted (c-index of 0.673 in the validation cohort), which is common in this kind of series with relatively uniform survival. These data reveal the intrinsic complexity of developing prognostic models within the context of AOA, where events are often stochastic and unpredictable and tend to cluster around the median survival time. Moreover, the validity of this nomogram has been verified only in a series of patients in which the HER2 testing positivity involved the use of anti-HER2 therapy. Despite this, it is worth noting that our model is well-calibrated and accurately predicts individual outcomes in both cohorts. Finally, the proposed nomogram requires external validation by means of prospectively collected data from independent cohorts of AOA patients undergoing first-line CT. In addition, in the era of molecular characterisation of gastric cancer, molecular subtypes associated with clinical phenotypes and different clinical outcomes have been delineated, thereby laying the groundwork for improved therapeutic strategies through the development of personalised medicine (Cristescu et al, 2015). Molecular markers other than HER2 that predict survival can therefore underpin clinical methods, such as this nomogram, to generate combined molecular and clinical models that refine individual risk prediction. However, to better understand the real impact of these complex and sophisticated molecular classifications it is increasingly important to determine the extent to which they provide additional discrimination ability over the information obtained through clinical scores, which are much easier to obtain and composed of routinely accessible factors.
In conclusion, we have developed and validated a new nomogram and a web-based calculator to predict OS in Caucasian AOA patients undergoing first-line combination CT in the era of trastuzumab. This prognostic tool can assist clinicians in day-to-day decision-making, such as risk stratification, individualised therapy or active treatment limitation determinations, as well as to prospectively categorise patients in forthcoming clinical trials.
Change history
06 June 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Rüschoff J, Kang YK (2010) Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376 (9742): 687–697.
Carmona-Bayonas A, Jiménez-Fonseca P, Lorenzo MLS, Ramchandani A, Martínez EA, Custodio A, Garrido M, Echavarría I, Cano JM, Barreto JEL, García TG, Manceñido FÁ, Lacalle A, Cardona MF, Mangas M, Visa L, Buxó E, Azkarate A, Díaz-Serrano A, Montes AF, Rivera F (2016) On the effect of triplet or doublet chemotherapy in advanced gastric cancer: results from a National Cancer Registry. J Natl Compr Canc Netw 14 (11): 1379–1388.
Charlson M, Szatrowski TP, Peterson J, Gold J (1994) Validation of a combined comorbidity index. J Clin Epidemiol 47 (11): 1245–1251.
Chau I, Norman AR, Cunningham D, Waters JS, Oates J, Ross PJ (2004) Multivariate prognostic factor analysis in locally advanced and metastatic esophago-gastric cancer—pooled analysis from three multicenter, randomized, controlled trials using individual patient data. J Clin Oncol 22 (12): 2395–2403.
Chau I, Ashley S, Cunningham D (2009) Validation of the Royal Marsden hospital prognostic index in advanced esophagogastric cancer using individual patient data from the REAL 2 study. J Clin Oncol 27 (19): e3–e4.
Cristescu R, Lee J, Nebozhyn M, Kim K-M, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K (2015) Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 21 (5): 449–456.
Custodio A, Madero R, Feliu J, Echevarría I, Hernández R, Cano J, Mangas M, Sánchez Lorenzo M, Jiménez-Fonseca P, Carmona-Bayonas A (2016) Desarrollo y validación de un modelo predictivo de supervivencia global (SG) en pacientes con cáncer gástrico avanzado (CGA) tratados con poliquimioterapia en primera línea. Spanish Soc Med Oncol Symp 74.
Duma N, Sanchez LJ, Castro YS, Jennis AA, McCain DA, Gutierrez ME, Bamboat ZM (2016) Gastric adenocarcinoma: clinicopathologic differences among Hispanics and non-Hispanic whites. A single Institution’s experience over 14 years. Ann Gastroenterol 29 (3): 325–331.
European Association for the Study of the Liver (2010) EASL clinical practice guidelines on the managament of ascites, spontaneous bacterial peritonitis, and hepatorenal syndrome in cirrhosis. J Hepatol 53 (3): 397–416.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136 (5): E359–E386.
Grenader T, Waddell T, Peckitt C, Oates J, Starling N, Cunningham D, Bridgewater J (2016) Prognostic value of neutrophil to lymphocyte ratio in advanced oesophago-gastric cancer: exploratory analysis of the REAL-2 trial. Ann Oncol 27 (4): 687–692.
Harrell FE (2015) Regression Modeling Strategies: with Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis 2nd edn Springer: New York, NY, USA.
Harrell F Jr, Frank E, Maintaner Frank E (2015) ‘Package ’rms‘.’ Vanderbilt University, Nashville, USA. Available from http://cran.r-project.org/web/packages/rms/index.html.
Hsieh M-C, Wang S-H, Chuah S-K, Lin Y-H, Lan J, Rau K-M (2016) A prognostic model using inflammation-and nutrition-based scores in patients with metastatic gastric adenocarcinoma treated with chemotherapy. Medicine 95 (17): e3504.
Jiménez-Fonseca P, Carmona-Bayonas A, Lorenzo MLS, Plazas JG, Custodio A, Hernández R, Garrido M, García T, Echavarría I, Cano JM (2017) Prognostic significance of performing universal HER2 testing in cases of advanced gastric cancer. Gastric Cancer 20 (3): 465–474.
Kang L, Chen W, Petrick NA, Gallas BD (2015) Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med 34 (4): 685–703.
Kawakami T, Narita Y, Oze I, Kadowaki S, Machida N, Taniguchi H, Ura T, Ando M, Ito S, Yatabe Y, Tajika M, Kito Y, Haraguchi Y, Yasui H, Muro K (2016) Establishment and validation of prognostic nomograms including HER2 status in metastastatic gastric cancer. J Clin Oncol 34 (suppl 4): 24.
Kim JG, Ryoo B-Y, Park YH, Kim B-S, Kim T-Y, Im Y-H, Kang Y-K (2008) Prognostic factors for survival of patients with advanced gastric cancer treated with cisplatin-based chemotherapy. Cancer Chemother Pharmacol 61 (2): 301–307.
Lee J, Lim T, Uhm JE, Park KW, Park SH, Lee SC, Park JO, Park YS, Lim HY, Sohn TS (2007) Prognostic model to predict survival following first-line chemotherapy in patients with metastatic gastric adenocarcinoma. Ann Oncol 18 (5): 886–891.
Maltoni M, Caraceni A, Brunelli C, Broeckaert B, Christakis N, Eychmueller S, Glare P, Nabal M, Vigano A, Larkin P (2005) Prognostic factors in advanced cancer patients: evidence-based clinical recommendations—a study by the Steering Committee of the European Association for Palliative Care. J Clin Oncol 23 (25): 6240–6248.
Ock CY, Nam AR, Lee J, Bang JH, Lee KH, Han SW, Kim TY, Im SA, Kim TY, Bang YJ, Oh DY (2016) Prognostic implication of antitumor immunity measured by the neutrophil-lymphocyte ratio and serum cytokines and angiogenic factors in gastric cancer. Gastric Cancer 20 (2): 254–262.
Ohtsu A, Shah MA, Van Cutsem E, Rha SY, Sawaki A, Park SR, Lim HY, Yamada Y, Wu J, Langer B (2011) Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a randomized, double-blind, placebo-controlled phase III study. J Clin Oncol 29 (30): 3968–3976.
Sun LB, Zhao GJ, Ding DY, Song B, Hou RZ, Li YC (2014) Comparison between better and poorly differentiated locally advanced gastric cancer in preoperative chemotherapy: a retrospective, comparative study at a single tertiary care institute. World J Surg Oncol 12: 280.
Takahari D, Boku N, Mizusawa J, Takashima A, Yamada Y, Yoshino T, Yamazaki K, Koizumi W, Fukase K, Yamaguchi K, Goto M, Nishina T, Tamura T, Tsuji A, Ohtsu A (2014) Determination of prognostic factors in Japanese patients with advanced gastric cancer using the data from a randomized controlled trial, Japan clinical oncology group 9912. Oncologist 19 (4): 358–366.
Takahari D, Mizusawa J, Koizumi W, Hyodo I, Boku N (2017) Validation of the JCOG prognostic index in advanced gastric cancer using individual patient data from the SPIRITS and G-SOX trials. Gastric Cancer e-pub ahead of print 16 February 2017 doi:10.1007/s10120-017-0702-0.
Vergouwe Y, Steyerberg EW, Eijkemans MJ, Habberma JD (2005) Substantial effective sample sizes were required for external validation studies of predictive logistic regression models. J Clin Epidemiol 58 (5): 475–483.
Waddell T, Chau I, Cunningham D, Gonzalez D, Okines AFC, Wotherspoon A, Saffery C, Middleton G, Wadsley J, Ferry D (2013) Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol 14 (6): 481–489.
Wagner A, Grothe W, Haerting J, Kleber G, Grothey A, Fleig WE (2006) Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J Clin Oncol 24 (18): 2903–2909.
Wang J, Qu J, Li Z, Che X, Zhang J, Liu J, Teng Y, Jin B, Zhao M, Liu Y (2016) A prognostic model in metastatic or recurrent gastric cancer patients with good performance status who received first-line chemotherapy. Transl Oncol 9 (3): 256–261.
Acknowledgements
We wish to thank the investigators of the AGAMENON study; Natalia Cateriano, Miguel Vaquero and IRICOM S.A. for the support of the website registry, and Priscilla Chase Duran for editing the manuscript.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Ethical statement
The study has been performed in accordance with the ethical standards of the Declaration of Helsinki and its later amendments. This study is an observational trial without intervention.
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Custodio, A., Carmona-Bayonas, A., Jiménez-Fonseca, P. et al. Nomogram-based prediction of survival in patients with advanced oesophagogastric adenocarcinoma receiving first-line chemotherapy: a multicenter prospective study in the era of trastuzumab. Br J Cancer 116, 1526–1535 (2017). https://doi.org/10.1038/bjc.2017.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.122
Keywords
This article is cited by
-
Efficacy and safety of chemotherapy in young patients with advanced gastroesophageal adenocarcinoma: data from the Spanish AGAMENON-SEOM registry
Gastric Cancer (2024)
-
Is there a preferred platinum and fluoropyrimidine regimen for advanced HER2-negative esophagogastric adenocarcinoma? Insights from 1293 patients in AGAMENON–SEOM registry
Clinical and Translational Oncology (2024)
-
Does HER2 status influence in the benefit of ramucirumab and paclitaxel as second line treatment of advanced gastro-esophageal adenocarcinoma? Data from the AGAMENON-SEOM registry
Journal of Cancer Research and Clinical Oncology (2023)
-
Development and validation of a prognostic scoring model for mortality risk stratification in patients with recurrent or metastatic gastric carcinoma
BMC Cancer (2021)
-
Prediction of quality of life in early breast cancer upon completion of adjuvant chemotherapy
npj Breast Cancer (2021)