Abstract
The ionotropic glutamate delta receptor GluD1, encoded by the GRID1 gene, is involved in synapse formation, function, and plasticity. GluD1 does not bind glutamate, but instead cerebellin and D-serine, which allow the formation of trans-synaptic bridges, and trigger transmembrane signaling. Despite wide expression in the nervous system, pathogenic GRID1 variants have not been characterized in humans so far. We report homozygous missense GRID1 variants in five individuals from two unrelated consanguineous families presenting with intellectual disability and spastic paraplegia, without (p.Thr752Met) or with (p.Arg161His) diagnosis of glaucoma, a threefold phenotypic association whose genetic bases had not been elucidated previously. Molecular modeling and electrophysiological recordings indicated that Arg161His and Thr752Met mutations alter the hinge between GluD1 cerebellin and D-serine binding domains and the function of this latter domain, respectively. Expression, trafficking, physical interaction with metabotropic glutamate receptor mGlu1, and cerebellin binding of GluD1 mutants were not conspicuously altered. Conversely, upon expression in neurons of dissociated or organotypic slice cultures, we found that both GluD1 mutants hampered metabotropic glutamate receptor mGlu1/5 signaling via Ca2+ and the ERK pathway and impaired dendrite morphology and excitatory synapse density. These results show that the clinical phenotypes are distinct entities segregating in the families as an autosomal recessive trait, and caused by pathophysiological effects of GluD1 mutants involving metabotropic glutamate receptor signaling and neuronal connectivity. Our findings unravel the importance of GluD1 receptor signaling in sensory, cognitive and motor functions of the human nervous system.
Similar content being viewed by others
Introduction
Intellectual disability (ID) and spastic paraplegia (SPG) are central nervous system disorders with marked clinical and genetic heterogeneity [1, 2]. The association of SPG with ID or MR (mental retardation, the out of use designation of ID) is frequent with 106 and 127 entries in the OMIM (Online Mendelian Inheritance in Man) database, respectively. Conversely, the triple combination of ID, SPG and glaucoma appears only once (OMIM#278050) with description of four patients of both sexes in two sibships of a large inbred pedigree [3], and of three male siblings born to first-cousin parents [4]. Although the consanguinity and presence of affected females suggest an autosomal recessive inheritance, the genetic basis of this distinct entity is unknown.
Glutamate delta receptors GluD1 (encoded by the GRID1 gene) and GluD2 (GRID2 gene) belong to the family of ionotropic glutamate receptors (iGluRs), which consist in homo- or heterotetrameric arrangements of subunits, and play key roles in synaptic transmission and plasticity [5,6,7]. GluDs do not bind glutamate but, instead, the binding of cerebellin and D-serine on distinct extracellular domains cooperatively gate GluD ion channels, whose opening is alternatively triggered by activation of Gq-coupled metabotropic glutamate receptors (mGlu1/5), or α1-adrenergic receptors [8,9,10,11]. The binding of these ligands also triggers or modulates metabotropic signals, cerebellin additionally enabling postsynaptic GluDs to participate in excitatory synapse formation/stabilization via attachment with presynaptic neurexin [6, 7, 12,13,14]. GluD1 and GluD2 are widely expressed in the brain at excitatory postsynaptic sites, GluD1 predominating over GluD2 outside the cerebellum [15,16,17]. The implication of GRID1 in pathology is suggested by association of GRID1 variants with risk of neuropsychiatric disorders [18,19,20,21,22,23,24,25,26], and by alterations observed in Grid1−/− mice at behavioral, cognitive, synaptic, and mGlu1/5 signaling levels [13, 27,28,29,30,31]. Yet, truly pathogenic GRID1 variants have not been described in human disease so far.
Here, we report the identification of homozygous missense variants in the GRID1 gene by genome-wide linkage analysis and/or whole exome sequencing (WES) in siblings from two unrelated consanguineous families presenting with mild or moderate ID, non- or slowly-progressive SPG, with (p.Arg161His) or without (p.Thr752Met) diagnosis of open angle glaucoma. Molecular modeling and experimental studies indicated that the mutations alter structural interactions within GluD1 extracellular domains, impact the D-serine binding site, and impair GluD1 effects on mGlu1/5 signaling, dendrite morphology, and excitatory synapse density in rodent neurons.
Materials and methods
Detailed materials and methods are provided as Supplementary Information
Patients
Written informed consent for genetic analysis was obtained from all participants or their legal guardians according to the Declaration of Helsinki and following Institutional Review Board (IRB)-approved protocols in the Centre Hospitalier Universitaire de Tours (Family A) and the Hadassah Medical Center (Family B).
Animals
Animal breeding and euthanasia were performed in accordance to European Communities Council Directive 86/609/062. Grid1 KO mice [32] (gift from Jian Zuo, Memphis, TE, USA) and Grid1−/− embryos obtained from heterozygous parents were genotyped as described [16]. Wild-type (WT) mice were purchased from Janvier Labs. All mice had C57BL/6 background.
Genome wide-linkage analysis and whole exome sequencing
Genotyping of Family A was performed on Genechip® human 250 K NspI array (Affymetrix) according to manufacturer’s instructions. Whole exome sequencing (WES) was performed using SureSelect Human All Exon kit (Agilent technologies) and the HiSEQ 2000 sequencer (Illumina). For Family B, DNA sample of the proband was shipped to Otogenetics, USA (CLIA lab). Sequencing data were aligned with the Human reference genome (hg19). Genetic segregation of the candidate variant with the disease was confirmed by Sanger sequencing.
Molecular modeling of GluD1 mutants structure
The protein was generated using Rat GluD1 receptor in complex with 7-chloro-kynurenate and Ca2+ as structure templates [33] (PDB codes: 6KSS and 6KSP), and prepared in the CHARMM-GUI web server [34] to insert protein in a membrane and solvate with water and ions. Mutant models were generated using Built Mutant protocol from Discovery Studio 2019. Molecular docking experiments of D-Serine, glycine and kynurenic acid at the active site were performed as described [35, 36]. Molecular dynamics were performed using NAMD protocols.
Plasmids and viruses
Plasmids encoding mouse WT GluD1 (GluD1WT) and GluD1 variants, rat mGlu1a, Green Fluorescent Protein (GFP), or tdTomato were used for transfection of HEK293 cells or neurons. Some constructs comprised a hemagglutinin (HA) epitope inserted after the predicted signal peptides of GluD1 and mGlu1a, this latter additionally comprising the Venus GFP variant fused to its C-terminus [9, 37]. Recombinant lentiviruses co-expressing GluD1 isoforms together with GFP were generated as described [9] and pseudo-virions produced at the Necker Institute Viral Vector and Gene Transfer facility (IFR94, Paris, France). Recombinant sindbis virus encoding the Ca2+ sensor Twitch-2B [38] was produced as described [39].
HEK293T cells
HEK293T cells (ATCC Number: CRL-3216) culture, plasmid transfection, membrane protein extraction, immunoprecipitation, western blotting and immunocytochemistry were performed using standard techniques and antibodies listed in Supplementary Table. For cerebellin binding experiments, HEK cells expressing GluD1 or GluD1 variants were incubated with 20 µg/ml recombinant human HA-tagged Cerebellin 1 (Cbln1, Biotechne 6934-CB-025) prior to rinsing and immunostaining.
Primary cortical or hippocampal cell cultures
Primary cortical or hippocampal cell cultures were prepared and cultured as described [40] from E17-E18 Grid1−/− or Grid1+/+ mice embryos, respectively. Western blotting and immunocytochemistry were performed using standard techniques and antibodies listed in Supplementary Table. For analyses of mGlu1/5 signaling, cortical cell cultures were transduced at 10 days in vitro (DIV) with GluD1-expressing lentiviruses, cultured for 4–7 additional days. Cultures were then either tested for ERK activity or transduced with Twitch-2B expressing sindbis virus the day before Ca2+ imaging experiments. ERK activity was measured on cultures incubated for 1 hour in medium containing 300 nM TTX (Latoxan) and 50 µM of the NMDAR antagonist APV (Hello Bio), then incubated in the presence or absence of RS-3,5-dihydroxyphenylglycine 2 (DHPG, 100 µM, Hello Bio), and finally processed for western blotting. Ca2+ imaging was performed in a perfusion chamber and responses to S-DHPG (50 µM, Hello Bio) monitored using a custom-built 2-photon laser scanning microscope as described [41]. For analyses of neurites and excitatory synapses, primary hippocampal cell cultures were transfected at DIV4 (dendritic morphometry) or DIV11-DIV13 (spines and synapses). Cultures were next incubated for 48 h, and then processed for immunocytochemistry.
Statistical analyses
All experiments were repeated at least three times. When d’Agostino-Pearson normality tests were successfully passed, we used One-way ANOVA parametric test, followed by Tukey’s post hoc method. For samples that did not pass the normality test, we used Kruskal–Wallis method followed by Dunn’s post hoc test. Results are given as mean ± standard error of the mean. Differences were considered significant if p < 0.05.
Results
Clinical description of the families
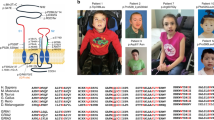
Family A included three affected brothers born to first-degree cousins (Fig. 1A), and presenting with non- or slowly-progressive SPG diagnosed in infancy with no other neurological signs, mild/moderate ID with normal occipitofrontal circumference, and juvenile open angle glaucoma causing severe visual impairment. This clinical picture is strikingly similar to earlier descriptions of this syndrome [3, 4]. Brain MRI, electromyography, metabolic investigations in one patient, and standard chromosome analysis of the three brothers were normal. Linkage to genes ARX, XNP, PLP and L1CAM was excluded, sequencing of MECP2 did not detect a causative variant, and high-resolution array-Comparative Genomic Hybridization (CGH) analysis did not reveal pathogenic copy number variation related to the disease.
A Pedigree of the families. Filled symbols indicate individuals with ID, SPG and Glaucoma (left), or ID and SPG (right). Individual number is indicated below each symbol. B Sanger sequencing electrophoregrams showing the GRID1 homozygous missense mutations c.482G>A, p.Arg161His and c.2255C>T, p.Thr752Met in the affected patients and the heterozygous mutations in unaffected relatives. C Amino-acid alignments showing conservation of GluD1 R161 and T752 residues across species, but not among iGluR family members. D Spatial organization of the transsynaptic complex GluD1-cerebellin (Cbln1)-neurexin at the glutamatergic synapse. Note that R161 and T752 residues belong to cerebellin-binding (ATD) and D-serine-binding domains (LBD), respectively.
Family B included two affected sisters, born to consanguineous parents (Fig. 1A). The proband presented at 6 years of age with global developmental delay, spastic paraplegia, craniosynostosis, dysmorphic features (brachycephaly, bilateral ptosis), and minor skeletal anomalies. Her 24-year old sister was similarly affected, but also showed kyphosis of the cervical spine, and required a wheelchair. Ophthalmologic examination could not be performed on either sister. The clinical features for Family B are detailed in Supplementary Information. Initial genetic investigations for the proband of Family B included chromosomal karyotype analysis which was normal, as well as CGH analysis, which was considered normal but notable for an intronic 50Kb deletion in 7q36.2 encompassing the DPP6 gene (arr:7q36.2(153,921,762-153,951,944)X1).
The clinical features for the five affected individuals are summarized in Table 1, together with side-by-side comparison with earlier observations [3, 4].
Identification of homozygous variants in GRID1
As consanguinity in both family pedigrees suggests autosomal recessive inheritance (Fig. 1A), we performed genome-wide single nucleotide polymorphism (SNP) genotyping for Family A in the three affected brothers, one healthy brother and both parents. Two homozygous regions with significant linkage were found at chromosome 10q23.1-q25.2 region (30.6 Mb between rs11201697 and rs7077757 markers, see Supplementary Fig. S1) and at chromosome 12q24.33 region (161 kb between rs10773690 and rs4759984 markers). WES analysis on two affected brothers and their father allowed the identification of a homozygous missense mutation of the GRID1 gene (NM_017551.2: c.482G>A, p.Arg161His; hg19, chr10:87966159C>T) within the 10q22q23 candidate region. This variant segregated in an autosomal recessive manner in all affected members of Family A (Fig. 1A, B, Table 1). The p.Arg161His GRID1 variant, predicted as “Disease causing” by Mutation Taster (score 0.9697) and referred in dbSNP (rs771100097), is only found at heterozygous state in 4 individuals from gnomAD database (Minor Allele Frequency, MAF = 1.60e−5).
For Family B, single (proband-only) WES was pursued, and brought to the identification of a homozygous missense variant in GRID1 (NM_017551.3: c.2255C>T, p.Thr752Met; hg19, chr10:87379729G>A). Using Sanger sequencing, this variant was confirmed to segregate with the disease in Family B, with both affected sisters homozygous for the variant, both parents and an unaffected male sibling found to be heterozygous carriers, and three additional unaffected siblings wild type for the variant (Fig. 1A, B, Table 1). The p.Thr752Met variant is only found at heterozygous state, in 11 individuals from gnomAD database (MAF = 3.89e−5).
Finally, sequencing of a cohort of more than 200 patients affected with SPG, isolated or associated with ID, failed to identify additional variants in GRID1.
Impact of Arg161His and Thr752Met mutations on GluD1 extracellular domains
The p.Arg161His (R161H) and p.Thr752Met (T752M) mutations concern GluD1 amino acid residues conserved among vertebrate species, but not among iGluR subunits (Fig. 1C), consistent with functional heterogeneity within this family [5, 33, 42]. Based on GluD1 3D structure [33], we assigned R161 and T752 residues to extracellular Amino Terminal Domain (ATD) and Ligand Binding Domain (LBD), which bind cerebellin and D-serine, respectively (Fig. 1D), the R161 residue being at the ATD-LBD interface distant from cerebellin binding residues, and the T752 residue lying within the LBD (Fig. 2A, C). We modeled the complete GluD1 structure by generating unresolved 3D loops crucial for activation [33] (Supplementary Fig. S2), and characterized effects of the mutations on this model. We found that the R161H mutation impacts the hinge between ATD and LBD by modifying the binding pattern with Q416, D417, and P419 residues of the loop linking the two domains, with possible consequences on their cooperativity [11, 33, 43]. In molecular dynamics experiments, the interaction patterns of H161 with Q416, E417 and P419 were not fully conserved over time (Fig. 2B), arguing for instability of ATD-LBD interaction in the mutant protein. The T752M mutation results in additional interactions between M752, Y748 (in the same α helix) and I729 (in adjacent α helix) that could lead to a stiffening of LBD. Further modeling experiments on the mutant protein showed a conservation of these interactions (i.e., M752 with Y748 and I729) over molecular dynamics duration and support a stiffening of the LBD. (Fig. 2D). Molecular docking indicated that D-serine affinity is decreased in GluD1T752M, but little modified in GluD1R161H (binding energy: wild-type GluD1 (GluD1WT), −107.9; GluD1R161H, −102.7; GluD1T752M, −68.8 kJ/mole), whereas binding of endogenous ligand glycine and synthetic ligand 7-chloro-kynurenate [44] are weakened by both mutations (from −100.7 and −108.8 kJ/mole in GluD1WT, to −76.8 and −79.1 kJ/mole in GluD1R161H, and to −65,7 and −77.8 kJ/mole in GluD1T752M, respectively). Electrophysiological analyses of the ion current mediated by a constitutively open channel GluD1 isoform [45] revealed that modulation of this current by D-serine and glycine was impaired by the T752M mutation, but moderately affected by the R161H mutation (Supplementary information and Supplementary Fig. S3), consistent with predictions of molecular modeling. These results indicate that R161H and T752M mutations can both affect GluD1 function by altering ligand binding and/or its transduction to transmembrane/intracellular signaling.
A, C Structure of the GluD1 homotetramer sitting above the plasma membrane - adapted from ref. [33]. Mutations affect residues situated at the interface (R161) between ATD and LBD extracellular domains, or within LBD (T752). B, D Predicted interactions of wt R161 and T752 residues, and of mutant H161 and M752 residues. The R161H mutation suppresses interaction with D417 residue of the loop linking ATD to LBD, thereby changing loop conformation (t = 0 ns). These interactions were also decreased during molecular dynamics (t = 10 ns), highlighting the weakness of ATD-LBD interactions in mutant protein. The T752M mutation results in supplementary interaction with I729 and Y748 residues of the LBD, thereby rigidifying this latter domain. These additional interactions, preserved during molecular dynamics, lead to a stiffening of LBD.
The R161H and T752M mutations do not hamper cerebellin binding to GluD1
To gain insight into the functional consequences of the mutations, we first expressed GluD1WT, GluD1R161H, and GluD1T752M in HEK cells, and found that GluD1 amount, molecular weight, and membrane insertion were not conspicuously affected by the mutations (Supplementary Fig. S4A). Transfection of hippocampal primary cell cultures confirmed plasma membrane expression of all variants in putative excitatory neurons with similar distribution along dendritic shafts and spines (Supplementary Fig. S4B). Hence, the pathogenicity of GluD1R161H and GluD1T752M does not result from deficits in their expression, stability or trafficking. We next compared the ability of GluD1WT, GluD1R161H and GluD1T752M to bind extracellular Cerebellin-1 in HEK cells, and found that both mutants retained the cerebellin-binding capability of GluD1WT (Supplementary Fig. S5). This was confirmed using Bio-Layer Interferometry measurements of interactions of recombinant Cerebellin-1 with WT or mutant GluD1 extracellular domains, which did not reveal differences in interaction kinetics or affinity that may hamper cerebellin binding to GluD1 mutants (Supplementary information and Supplementary Fig. S6). Hence, cerebellin binding, and thus trans-synaptic scaffolding ability [6, 12, 14], is essentially preserved in both mutants, consistent with R161H and T752M mutations being distant from cerebellin-binding residues in the GluD1 3D structure.
The R161H and T752M mutations impair the modulation of mGlu1/5 signaling by GluD1
Our above results suggest that R161H and T752M mutations can affect GluD1 function by altering ligand binding and its transduction to transmembrane/intracellular signaling [6, 7, 11, 14]. We thus searched for alteration of mGlu1/5 signaling, which involves GluD1, is impaired in Grid1−/− mice, and whose dysregulation at the level of non-canonical pathways tightly relates to ID and related neurodevelopmental disorders [9, 30, 46,47,48].
We first verified that mGlu1 co-immunoprecipitated with GluD1WT, GluD1R161H, or GluD1T752M with similar efficiency upon co-expression in HEK (Supplementary Fig. S7), indicating that the mGlu1-GluD1 physical interaction [9, 30] is not impaired by the R161H and T752M mutations.
Next, GluD1WT, GluD1R161H, or GluD1T752M were co-expressed with GFP through lentiviral transduction in primary cultures of cortical cells from Grid1−/− mice, which avoid influence of endogenous GluD1WT on mGlu1/5 signaling. All GFP-labeled transduced cells examined were GluD1-immunopositive (Fig. 3A), and the vast majority of neurons in these cultures were transduced (Supplementary Fig. S8). In a first set of experiments, cultures were incubated 5 min in the presence/absence of the mGlu1/5 agonist DHPG (100 µM), and then processed for western blot and immunoquantification of the phosphoERK/ERK ratio (Fig. 3B). In GluD1WT-expressing cultures, DHPG treatment strongly increased the phosphoERK/ERK ratio relative to mock-treated control cultures (DHPG: 221 ± 8% of control; n = 27 control, n = 27 DHPG-treated cultures, Fig. 3B). The same paradigm elicited a significantly smaller increase of phosphoERK/ERK ratio in GluD1R161H-expressing (DHPG: 161 ± 9% of control, n = 14 DHPG-GluD1R161H) or GluD1T752M-expressing cultures (DHPG: 180 ± 12% of control, n = 10 DHPG-GluD1T752M; Fig. 3B). In a second set of experiments, live imaging of Ca2+responses of individual neurons to S-DHPG (50 µM) was performed using the ratiometric fluorescent sensor Twitch-2B additionally expressed through Sindbis viral transfer. Drug application elicited a transient increase of Twitch-2B fluorescence ratio, indicative of intracellular Ca2+ increase. Changes of fluorescence ratio had a significantly larger peak amplitude in GluD1WT- (13 ± 1%, n = 63), than in GluD1R161H- (7 ± 1%, n = 31) and GluD1T752M– expressing neurons (9 ± 1%, n = 51). Moreover, Twitch-2B response integral was significantly reduced in GluD1R161H- and GluD1T752M–expressing neurons (to 57 ± 5 and 66 ± 5%, respectively) compared to GluD1WT-expressing neurons (Fig. 3C). These results indicate that the modulation by GluD1 of mGlu1/5 intracellular signaling via both Ca2+ and the ERK pathway is impaired by the R161H and T752M mutations.
A Fluorescence pictures of primary cortical cell cultures from Grid1−/− mouse co-expressing GluD1WT/GluD1R161H/GluD1T752M and GFP following lentiviral transfer, scale bar: 50 µm. B Western blot analysis of virally transduced cortical cultures following incubation in presence or absence of the mGlu1/5 agonist DHPG (100 µM) and graph summarizing results obtained in mock-treated or DHPG-treated cultures expressing GluD1WT (n = 27 and 27, respectively), GluD1R161H (n = 12 and 14, respectively), or GluD1T752M (n = 10 and 10, respectively). C 2-photon Ca2+ imaging in cortical cell cultures co-expressing GluD1WT/GluD1R161H/GluD1T752M and the ratiometric Twitch-2B sensor following dual lenti/sindbis viral transfer. Traces show mean (lines) ±SEM (shade) changes of Twitch-2B YFP/CFP ratio in response to S-DHPG (50 µM), indicative of transient Ca2+ increase in somata of neurons expressing GluD1WT (n = 63), GluD1R161H (n = 31), or GluD1T752M (n = 51). Graphs summarize results obtained from cells of at least 6 coverslips and 2 cultures per condition. *Significant differences.
The GluD1 R161H and T752M mutations impair dendrite morphology and excitatory synapse density
Alterations of dendrites and synapses are found in ID and related neurodevelopmental disorders in humans and mouse models [49,50,51]. We thus co-transfected plasmids encoding GluD1WT, GluD1R161H, or GluD1T752M together with a GFP-encoding plasmid in hippocampal primary cell cultures from Grid1+/+ mice and examined the morphology of GFP-expressing neurons. The vast majority of GFP-positive neurons also over-expressed either GluD1WT (96.8 ± 4.2%, n = 252), GluD1R161H (96.3 ± 3.9%, n = 246), or GluD1T752M (96.1 ± 4.7%, n = 250). Moreover, plasmid-driven expression of GluD1WT and mutants was largely superior to that of endogenous GluD1 (Supplementary Fig. S9), allowing the effects of recessive R161H and T752M mutations to be evaluated in transfected Grid1+/+ neurons.
Analyses of GFP-labeled neurites using the Sholl method revealed a significant reduction in total neuritic length of neurons overexpressing GluD1 mutants, as compared to control (GFP only) and GluD1WT-overexpressing neurons (control: 598 ± 33 µm, GluD1: 577 ± 25 µm, GluD1R161H: 466 ± 21 µm, GluD1T752M: 407 ± 21 µm; n = 41, 44, 42, 44 neurons, respectively, from 3 cultures in each condition; Fig. 4A). This was associated with a significantly reduced neuritic ramification of neurons transfected with GluD1R161H or GluD1T752M (total crossings; control: 184 ± 9, GluD1: 176 ± 9, GluD1R161H: 138 ± 6, GluD1T752M: 138 ± 6; n = 40, 45, 41, 44 neurons, respectively; Fig. 4A). These findings indicate that the GluD1 R161H and T752M mutations perturb neurite outgrowth and architecture.
A Binary images show GFP fluorescence of cultured hippocampal neurons expressing GFP alone, or GFP and indicated GluD1 variants, after plasmid transfection. Neurites crossing concentric circles centered on each neuron’s soma were counted to quantify neurite ramification. Total neurite length is the sum of all neuritic segments measured for each neuron. Graphs summarize results obtained in n ≥ 40 neurons from 3 cultures in each condition. B Squares on the GFP fluorescence picture of a pyramidal-shaped hippocampal neuron in transfected culture (upper left) exemplify regions where excitatory putative synapses, revealed by overlap of presynaptic Bassoon and postsynaptic Homer immunostaining on GFP positive dendrites (upper right), were counted. The graph shows results obtained in n ≥ 20 pyramidal-shaped hippocampal neurons from 3 cultures in each transfection condition indicated. *Significant differences.
GluD1 is present at excitatory postsynaptic sites [9, 15, 16], and promotes the formation of dendritic spines and excitatory synapses [12, 13, 29, 52]. We thus evaluated the effect of the R161H mutation on spine density and morphology, and found that this mutation impairs GluD1 stimulatory effects on dendritic spine formation and maturation (Supplementary information and Supplementary Fig. S10). Further analyses in hippocampal organotypic slice cultures indicated that R161H and T752M mutations also impair the enhancing effect of GluD1 on dendritic spine density in an integrated neural environment (Supplementary information and Supplementary Fig. S11). Finally, we examined the impact of the GluD1 R161H and T752M mutations on excitatory synapse density by counting overlaps of presynaptic Bassoon and postsynaptic Homer immunolabelling on GFP-expressing pyramidal-shaped neurons from hippocampal primary cell cultures. As shown in Fig. 4B, we observed a significantly higher density of putative excitatory synapses on both proximal and distal parts of apical and basal dendrites of GluD1WT-transfected compared to control neurons (apical proximal: 5.1 ± 0.4 vs. 2.5 ± 0.2; apical distal: 3.8 ± 0.3 vs. 2.0 ± 0.1; lateral proximal: 3.7 ± 0.2 vs. 2.0 ± 0.1; lateral distal: 3.4 ± 0.3 vs. 1.9 ± 0.1 per 10 µm dendrite of GluD1WT vs. control neurons, respectively; n = 25, 24, 26, 24 GluD1WT and n = 28, 27, 28, 27 control neurons, respectively), consistent with the synaptogenic function of GluD1 [12, 29, 52]. Conversely, the density of excitatory synapses on neurons overexpressing GluD1R161H or GluD1T752M was similar to that on control neurons (apical proximal: 2.6 ± 0.2 and 2.5 ± 0.1; apical distal: 2.2 ± 0.2 and 2.1 ± 0.1; lateral proximal: 2.4 ± 0.2 and 2.1 ± 0.1; lateral distal: 2.2 ± 0.2 and 2.0 ± 0.1 per 10 µm dendrite of GluD1R161H and GluD1T752M neurons, respectively; n = 20, 25, 26, 23 GluD1R161H and n = 33, 33, 31, 31 GluD1T752M neurons, respectively), suggesting that the role of GluD1 in excitatory synapse formation and stabilization is hampered by the R161H and T752M mutations.
These results indicate that regulation of neurite outgrowth, architecture, spine density and maturation, and excitatory synapse density are impaired by the GluD1 R161H and T752M mutations. Given the widespread distribution [15, 16] of GluD1, the R161H and T752M mutations are thus likely to affect critically the formation and function of brain networks.
Discussion
We report association between homozygous missense variants p.Arg161His and p.Thr752Met in the GRID1 gene, and disease phenotypes including ID, SPG, and glaucoma, in two different sibships born to consanguineous parents. Our experimental findings indicate that these GRID1 variants impair mGlu1/5 signaling, as well as dendritic morphology and excitatory synapse density, in mouse forebrain neurons.
Homozygous GRID1 variants causing ID and SPG with or without glaucoma
Here, we characterize pathogenic recessive GRID1 mutations linked to syndromic ID and SPG without or with glaucoma, a triple phenotypic association whose genetic bases had not been elucidated previously. The constant association of ID with SPG and glaucoma is rare, as only two affected families have been described [3, 4], but glaucoma has been mentioned in patients affected with SPG45 and SP75, two conditions usually comprising only ID and SPG [53, 54]. The two affected sibships reported herein share some phenotypic features and differ in others. Inter- and intra-familial phenotypic variability is well-described in inherited neurodevelopmental disorders [55] and hereditary SPG [56], and might explain some of these differences. Characterization of the full clinical spectrum of the present syndrome and elucidation of possible genotype-phenotype correlations thus await identification of additional affected individuals.
GluD1 is a postsynaptic protein widely expressed in the brain [15, 16]. While many genetic variants linked to ID concern synaptic proteins [47,48,49], only few SPG-linked genes [2, 57], and no glaucoma-associated genes [58, 59] identified so far encode synaptic proteins. However, several SPG-linked variants impact synapses (e.g. AP4M1 variants altering iGluRs trafficking [60,61,62]), and synaptic changes appear to underlie early dysfunction of retinal ganglion cells in glaucoma [63]. Hence, the GRID1 p.Arg161His and p.Thr752Met variants are rare examples of genetic alteration in a synaptic protein causing ID and SPG with or without glaucoma, but the existence of such mutations is consistent with synaptic impairments occurring in all three pathologies.
The GluD1 R161H and T752M mutants impair mGlu1-5 signaling, dendrite architecture and excitatory synapses
We found that mGlu1/5 signaling via Ca2+ and the non-canonical ERK pathway is hampered by the GluD1 R161H and T752M mutations. Both Ca2+ and ERK signals are involved in neurite growth and maintenance, synapse formation and plasticity [64,65,66,67,68]. This suggests that part of the pathogenic impact of GluD1 R161H and T752M mutations stems from impaired signaling of the mGlu1/5-GluD1 complex. Nonetheless, additional deleterious effects of these mutations may occur due to dysregulation of yet other signaling mechanisms [10, 11, 14, 30] involving GluD1.
We also found that dendrite outgrowth, architecture, spine density and maturation, and excitatory synapse density are impaired by the GluD1 R161H and T752M mutations, despite cerebellin binding, thus trans-synaptic scaffolding, being preserved in both GluD1 mutants. This confirms that transmembrane signaling by GluD1 is essential to its role in the formation and regulation of excitatory synapses [12, 14]. Since loss of expression or function of GluD1 impairs diverse glutamatergic synapses in the forebrain, midbrain and cerebellum [9, 12,13,14,15, 29, 31], it is likely that the pathogenic effects of GluD1 R161H and T752M mutations on neuronal morphology and connectivity extend broadly in the brain.
Implications for ID, SPG and glaucoma
GluD1 is expressed in neurons that form connections altered in ID: neurons of forebrain networks [15, 16], in SPG: pyramidal cells of the motor cortex [15, 16] and their spinal motoneuron targets [69], and in glaucoma: retinal ganglion cells [70, 71] and their targets in sensory thalamus and superior colliculus [15, 16]. This indicates that the observed cellular impact of GluD1 mutants has direct relevance to these pathologies. Indeed, dysregulation of mGlu1/5 signaling and synaptic alterations in forebrain neurons are tightly linked to ID and related neurodevelopmental disorders [46,47,48,49,50,51]. Likewise, GluD1 mutants are likely to affect the formation and maintenance of long-range pyramidal tract and optic nerve connections by altering both projection and target neurons properties. The high sensitivity of the corticospinal tract to changes in ERK signaling level [72], and the importance of mGlu1/5 for the excitability of retinal ganglion cells and their connectivity to thalamic targets [73, 74], are documented indications that GluD1 mutants can indeed contribute to corticospinal axons and optic nerve damage that cause SPG and glaucoma, respectively.
In conclusion, we report the first pathogenic variants of the GRID1 gene in patients presenting with ID and SPG with or without glaucoma and provide evidence that these variants impair mGlu1/5 signaling, dendrite outgrowth, architecture, spine density and maturation, and synapse density. Although the present study does not exhaust the possible pathophysiological effects of GluD1R161H and GluD1T752M mutants, our observations demonstrate that their expression has deleterious consequences on neurons and circuits that can cause ID, SPG and glaucoma.
References
Ellison JW, Rosenfeld JA, Shaffer LG. Genetic basis of intellectual disability. Annu Rev Med. 2013;64:441–50.
Elsayed LEO, Eltazi IZ, Ahmed AE, Stevanin G. Insights into Clinical, Genetic, and Pathological Aspects of Hereditary Spastic Paraplegias: A Comprehensive Overview. Front Mol Biosci. 2021;8:690899.
Heijbel J, Jagell S. Spastic paraplegia, glaucoma and mental retardation-in three siblings. A new genetic syndrome. Hereditas. 1981;94:203–7.
Chenevix-Trench G, Leshner R, Mamunes P. Spastic paresis, glaucoma and mental retardation-a probable autosomal recessive syndrome? Clin Genet. 1986;30:416–21.
Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharm Rev. 2010;62:405–96.
Yuzaki M, Aricescu AR. A GluD Coming-Of-Age Story. Trends Neurosci. 2017;40:138–50.
Burada AP, Vinnakota R, Lambolez B, Tricoire L, Kumar J. Structural biology of ionotropic glutamate delta receptors and their crosstalk with metabotropic glutamate receptors. Neuropharmacology. 2021;196:108683.
Ady V, Perroy J, Tricoire L, Piochon C, Dadak S, Chen X, et al. Type 1 metabotropic glutamate receptors (mGlu1) trigger the gating of GluD2 delta glutamate receptors. EMBO Rep. 2014;15:103–9.
Benamer N, Marti F, Lujan R, Hepp R, Aubier TG, Dupin AAM, et al. GluD1, linked to schizophrenia, controls the burst firing of dopamine neurons. Mol Psychiatry. 2018;23:691–700.
Gantz SC, Moussawi K, Hake HS. Delta glutamate receptor conductance drives excitation of mouse dorsal raphe neurons. Elife. 2020;9:e56054.
Carrillo E, Gonzalez CU, Berka V, Jayaraman V. Delta glutamate receptors are functional glycine- and D-serine-gated cation channels in situ. Sci Adv. 2021;7:eabk2200.
Tao W, Díaz-Alonso J, Sheng N, Nicoll RA. Postsynaptic δ1 glutamate receptor assembles and maintains hippocampal synapses via Cbln2 and neurexin. Proc Natl Acad Sci USA. 2018;115:E5373–E5381.
Andrews PC, Dravid SM. An emerging map of glutamate delta 1 receptors in the forebrain. Neuropharmacology. 2021;192:108587.
Dai J, Patzke C, Liakath-Ali K, Seigneur E, Südhof TC. GluD1 is a signal transduction device disguised as an ionotropic receptor. Nature. 2021;595:261–5.
Konno K, Matsuda K, Nakamoto C, Uchigashima M, Miyazaki T, Yamasaki M, et al. Enriched expression of GluD1 in higher brain regions and its involvement in parallel fiber-interneuron synapse formation in the cerebellum. J Neurosci. 2014;34:7412–24.
Hepp R, Hay YA, Aguado C, Lujan R, Dauphinot L, Potier MC, et al. Glutamate receptors of the delta family are widely expressed in the adult brain. Brain Struct Funct. 2015;220:2797–815.
Nakamoto C, Konno K, Miyazaki T, Nakatsukasa E, Natsume R, Abe M, et al. Expression mapping, quantification, and complex formation of GluD1 and GluD2 glutamate receptors in adult mouse brain. J Comp Neurol. 2020;528:1003–27.
Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–36.
Guo S-Z, Huang K, Shi Y-Y, Tang W, Zhou J, Feng G-Y, et al. A case-control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr Res. 2007;93:385–90.
Treutlein J, Mühleisen TW, Frank J, Mattheisen M, Herms S, Ludwig KU, et al. Dissection of phenotype reveals possible association between schizophrenia and Glutamate Receptor Delta 1 (GRID1) gene promoter. Schizophr Res. 2009;111:123–30.
Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–73.
Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46.
Nenadic I, Maitra R, Scherpiet S, Gaser C, Schultz CC, Schachtzabel C, et al. Glutamate receptor δ 1 (GRID1) genetic variation and brain structure in schizophrenia. J Psychiatr Res. 2012;46:1531–9.
Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 2012;21:3513–23.
Fitzgerald TW, Gerety SS, Jones WD, van Kogelenberg M, King DA, McRae J, et al. Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–8.
Zhang T, Hou L, Chen DT, McMahon FJ, Wang JC, Rice JP. Exome sequencing of a large family identifies potential candidate genes contributing risk to bipolar disorder. Gene. 2018;645:119–23.
Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ, Dravid SM. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PloS One. 2012;7:e32969.
Yadav R, Hillman BG, Gupta SC, Suryavanshi P, Bhatt JM, Pavuluri R, et al. Deletion of glutamate delta-1 receptor in mouse leads to enhanced working memory and deficit in fear conditioning. PloS One. 2013;8:e60785.
Gupta SC, Yadav R, Pavuluri R, Morley BJ, Stairs DJ, Dravid SM. Essential role of GluD1 in dendritic spine development and GluN2B to GluN2A NMDAR subunit switch in the cortex and hippocampus reveals ability of GluN2B inhibition in correcting hyperconnectivity. Neuropharmacology. 2015;93:274–84.
Suryavanshi PS, Gupta SC, Yadav R, Kesherwani V, Liu J, Dravid SM. Glutamate Delta-1 Receptor Regulates Metabotropic Glutamate Receptor 5 Signaling in the Hippocampus. Mol Pharm. 2016;90:96–105.
Liu J, Shelkar GP, Gandhi PJ, Gawande DY, Hoover A, Villalba RM, et al. Striatal glutamate delta-1 receptor regulates behavioral flexibility and thalamostriatal connectivity. Neurobiol Dis. 2020;137:104746.
Gao J, Maison SF, Wu X, Hirose K, Jones SM, Bayazitov I, et al. Orphan glutamate receptor delta1 subunit required for high-frequency hearing. Mol Cell Biol. 2007;27:4500–12.
Burada AP, Vinnakota R, Kumar J. Cryo-EM structures of the ionotropic glutamate receptor GluD1 reveal a non-swapped architecture. Nat Struct Mol Biol. 2020;27:84–91.
Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J Comput Chem. 2008;29:1859–65.
Ducassou L, Jonasson G, Dhers L, Pietrancosta N, Ramassamy B, Xu-Li Y, et al. Expression in yeast, new substrates, and construction of a first 3D model of human orphan cytochrome P450 2U1: Interpretation of substrate hydroxylation regioselectivity from docking studies. Biochim Biophys Acta. 2015;1850:1426–37.
Dhers L, Pietrancosta N, Ducassou L, Ramassamy B, Dairou J, Jaouen M, et al. Spectral and 3D model studies of the interaction of orphan human cytochrome P450 2U1 with substrates and ligands. Biochim Biophys Acta Gen Subj. 2017;1861:3144–53.
Perroy J, Raynaud F, Homburger V, Rousset MC, Telley L, Bockaert J, et al. Direct interaction enables cross-talk between ionotropic and group I metabotropic glutamate receptors. J Biol Chem. 2008;283:6799–805.
Thestrup T, Litzlbauer J, Bartholomäus I, Mues M, Russo L, Dana H, et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat Methods. 2014;11:175–82.
Gervasi N, Hepp R, Tricoire L, Zhang J, Lambolez B, Paupardin-Tritsch D, et al. Dynamics of protein kinase A signaling at the membrane, in the cytosol, and in the nucleus of neurons in mouse brain slices. J Neurosci. 2007;27:2744–50.
Ung DC, Iacono G, Méziane H, Blanchard E, Papon MA, Selten M, et al. Ptchd1 deficiency induces excitatory synaptic and cognitive dysfunctions in mouse. Mol Psychiatry. 2018;23:1356–67.
Bonnot A, Guiot E, Hepp R, Cavellini L, Tricoire L, Lambolez B. Single-fluorophore biosensors based on conformation-sensitive GFP variants. FASEB J. 2014;28:1375–85.
Schmid SM, Hollmann M. To gate or not to gate: are the delta subunits in the glutamate receptor family functional ion channels? Mol Neurobiol. 2008;37:126–41.
Elegheert J, Kakegawa W, Clay JE, Shanks NF, Behiels E, Matsuda K, et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science. 2016;353:295–9.
Kristensen AS, Hansen KB, Naur P, Olsen L, Kurtkaya NL, Dravid SM, et al. Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors. Mol Pharm. 2016;89:253–62.
Yadav R, Rimerman R, Scofield MA, Dravid SM. Mutations in the transmembrane domain M3 generate spontaneously open orphan glutamate δ1 receptor. Brain Res. 2011;1382:1–8.
D’Antoni S, Spatuzza M, Bonaccorso CM, Musumeci SA, Ciranna L, Nicoletti F, et al. Dysregulation of group-I metabotropic glutamate (mGlu) receptor mediated signalling in disorders associated with Intellectual Disability and Autism. Neurosci Biobehav Rev. 2014;46:228–41.
Stoppel LJ, Auerbach BD, Senter RK, Preza AR, Lefkowitz RJ, Bear MF. beta-Arrestin2 Couples Metabotropic Glutamate Receptor 5 to Neuronal Protein Synthesis and Is a Potential Target to Treat Fragile X. Cell Rep. 2017;18:2807–14.
Wilkerson JR, Albanesi JP, Huber KM. Roles for Arc in metabotropic glutamate receptor-dependent LTD and synapse elimination: Implications in health and disease. Semin Cell Dev Biol. 2018;77:51–62.
Bagni C, Zukin RS. A Synaptic Perspective of Fragile X Syndrome and Autism Spectrum Disorders. Neuron. 2019;101:1070–88.
Banerjee A, Miller MT, Li K, Sur M, Kaufmann WE. Towards a better diagnosis and treatment of Rett syndrome: a model synaptic disorder. Brain. 2019;142:239–48.
Lima Caldeira G, Peça J, Carvalho AL. New insights on synaptic dysfunction in neuropsychiatric disorders. Curr Opin Neurobiol. 2019;57:62–70.
Ryu K, Yokoyama M, Yamashita M, Hirano T. Induction of excitatory and inhibitory presynaptic differentiation by GluD1. Biochem Biophys Res Commun. 2012;417:157–61.
Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–11.
Lossos A, Elazar N, Lerer I, Schueler-Furman O, Fellig Y, Glick B, et al. Myelin-associated glycoprotein gene mutation causes Pelizaeus-Merzbacher disease-like disorder. Brain. 2015;138:2521–36.
Hanly C, Shah H, Au PYB, Murias K. Description of neurodevelopmental phenotypes associated with 10 genetic neurodevelopmental disorders: A scoping review. Clin Genet. 2021;99:335–46.
Klebe S, Stevanin G, Depienne C. Clinical and genetic heterogeneity in hereditary spastic paraplegias: from SPG1 to SPG72 and still counting. Rev Neurol. 2015;171:505–30.
Blackstone C. Converging cellular themes for the hereditary spastic paraplegias. Curr Opin Neurobiol. 2018;51:139–46.
Liu Y, Allingham RR. Major review: Molecular genetics of primary open-angle glaucoma. Exp Eye Res. 2017;160:62–84.
Trivli A, Zervou MI, Goulielmos GN, Spandidos DA, Detorakis ET. Primary open angle glaucoma genetics: The common variants and their clinical associations. Mol Med Rep. 2020;22:1103–10.
Matsuda S, Miura E, Matsuda K, Kakegawa W, Kohda K, Watanabe M, et al. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57:730–45.
Abou Jamra R, Philippe O, Raas-Rothschild A, Eck SH, Graf E, Buchert R, et al. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am J Hum Genet. 2011;88:788–95.
Bettencourt C, Salpietro V, Efthymiou S, Chelban V, Hughes D, Pittman AM, et al. Genotype-phenotype correlations and expansion of the molecular spectrum of AP4M1-related hereditary spastic paraplegia. Orphanet J Rare Dis. 2017;12:172.
Agostinone J, Di Polo A. Retinal ganglion cell dendrite pathology and synapse loss: Implications for glaucoma. Prog Brain Res. 2015;220:199–216.
Lohmann C, Wong RO. Regulation of dendritic growth and plasticity by local and global calcium dynamics. Cell Calcium 2005;37:403–9.
Michaelsen K, Lohmann C. Calcium dynamics at developing synapses: mechanisms and functions. Eur J Neurosci. 2010;32:218–23.
Impey S, Obrietan K, Storm DR. Making new connections: role of ERK/MAP kinase signaling in neuronal plasticity. Neuron. 1999;23:11–14.
Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. J Neurochem. 2001;76:1–10.
Polleux F, Snider W. Initiating and growing an axon. Cold Spring Harb Perspect Biol. 2010;2:a001925.
Tölle TR, Berthele A, Zieglgänsberger W, Seeburg PH, Wisden W. The differential expression of 16 NMDA and non-NMDA receptor subunits in the rat spinal cord and in periaqueductal gray. J Neurosci. 1993;13:5009–28.
Brandstätter JH, Koulen P, Wässle H. Selective synaptic distribution of kainate receptor subunits in the two plexiform layers of the rat retina. J Neurosci. 1997;17:9298–307.
Jakobs TC, Ben Y, Masland RH. Expression of mRNA for glutamate receptor subunits distinguishes the major classes of retinal neurons, but is less specific for individual cell types. Mol Vis. 2007;13:933–48.
Xing L, Larsen RS, Bjorklund GR, Li X, Wu Y, Philpot BD, et al. Layer specific and general requirements for ERK/MAPK signaling in the developing neocortex. Elife. 2016;5:e11123.
Narushima M, Uchigashima M, Yagasaki Y, Harada T, Nagumo Y, Uesaka N, et al. The Metabotropic Glutamate Receptor Subtype 1 Mediates Experience-Dependent Maintenance of Mature Synaptic Connectivity in the Visual Thalamus. Neuron. 2016;91:1097–109.
Li Q, Cui P, Miao Y, Gao F, Li XY, Qian WJ, et al. Activation of group I metabotropic glutamate receptors regulates the excitability of rat retinal ganglion cells by suppressing Kir and Ih. Brain Struct Funct. 2017;222:813–30.
Acknowledgements
The authors thank the patients and their families for their kind cooperation, Pr. Jamel Chelly and Nicolas Lebrun for discussions and advice, Agnès Rastetter for help in genetic analyses, the Bioinformatics Platform of Imagine Institute (linkage analyses), and the IBPS Cell Imaging and Animal Facilities. The study received support and funding from the INSERM (GOLD cross-cutting program on genomic variability, to FL), European Union FP7 Project GENCODYS (to FL, grant number 241995), Association pour le Développement de la Neurogénétique (to FL), Fondation Maladies Rares (to AT), the French Agency for Research (to LT, ANR-16-CE16-0014-01) and the Jerôme Lejeune foundation (to RH, grant numbers 1693 and 1959). JE was recipient of a Chaire d’Excellence Neurocampus grant and a European Research Council Starting Grant (number 850820, ‘SynLink’).
Author information
Authors and Affiliations
Contributions
Examined patients: BPS, ARR, OE, BAL, NH and AT. Performed experiments: DCU, EBB, BR, DT, AZ, AD, OE, BAL, MAP, SM, RAT, GS, LT, RH and FL. Performed molecular modeling: NP. Analyzed data: DCU, EBB, BR, DT, BPS, ARR, OE, BAL, JE, ML, GS, LT, AT, RH and FL. Supervised the work: BPS, ARR, JE, ML, MH, GS, AT, BL, LT, RH, FL. Wrote the paper: DCU, NP, BPS, JE, ML, BL, LT, AT, RH and FL.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ung, D.C., Pietrancosta, N., Badillo, E.B. et al. GRID1/GluD1 homozygous variants linked to intellectual disability and spastic paraplegia impair mGlu1/5 receptor signaling and excitatory synapses. Mol Psychiatry (2024). https://doi.org/10.1038/s41380-024-02469-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-024-02469-w