Abstract
Gut-associated lymphoid tissues (GALTs) comprise key intestinal immune inductive sites, including the Peyer’s patches of the small intestine and different types of isolated lymphoid follicle (ILF) found along the length of the gut. Our understanding of human GALT is limited due to a lack of protocols for their isolation. Here we describe a technique that, uniquely among intestinal cell isolation protocols, allows identification and isolation of all human GALT, as well as GALT-free intestinal lamina propria (LP). The technique involves the mechanical separation of intestinal mucosa from the submucosa, allowing the identification and isolation of submucosal ILF (SM-ILF), LP-embedded mucosal ILF (M-ILF) and LP free of contaminating lymphoid tissue. Individual SM-ILF, M-ILF and Peyer’s patch follicles can be subsequently digested for downstream cellular and molecular characterization. The technique, which takes 4–10 h, will be useful for researchers interested in intestinal immune development and function in health and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Agace, W. W. & Mccoy, K. D. Immunity review regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46, 532–548 (2017).
Mowat, A. M. & Agace, W. W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14, 667–685 (2014).
Trepel, F. Number and distribution of lymphocytes in man. A critical analysis. Klin. Wochenschr. 52, 511–515 (1974).
Spencer, J. & Sollid, L. M. The human intestinal B-cell response. Mucosal Immunol. 9, 1113–1124 (2016).
Spencer, J., Siu, J. H. Y. & Montorsi, L. Human intestinal lymphoid tissue in time and space. Mucosal Immunol. 12, 296–298 (2019).
Smillie, C. S. et al. Intra- and Inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019).
Martin, J. C. et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508.e20 (2019).
Fenton, T. M. et al. Immune profiling of human gut-associated lymphoid tissue identifies a role for isolated lymphoid follicles in priming of region-specific immunity. Immunity 52, 557–570.e6 (2020).
O’Leary, A. D. & Sweeney, E. C. Lymphoglandular complexes of the colon: structure and distribution. Histopathology 10, 267–283 (1986).
Moghaddami, M., Cummins, A. & Mayrhofer, G. Lymphocyte-filled villi: comparison with other lymphoid aggregations in the mucosa of the human small intestine. Gastroenterology 115, 1414–1425 (1998).
Senda, T. et al. Microanatomical dissection of human intestinal T-cell immunity reveals site-specific changes in gut-associated lymphoid tissues over life. Mucosal Immunol. 12, 378–389 (2019).
Fujimura, Y., Hosobe, M. & Kihara, T. Ultrastructural study of M cells from colonic lymphoid nodules obtained by colonoscopic biopsy. Dig. Dis. Sci. 37, 1089–1098 (1992).
Junker, Y. et al. Comparative analysis of mononuclear cells isolated from mucosal lymphoid follicles of the human ileum and colon. Clin. Exp. Immunol. 156, 232–237 (2009).
Meier, D. et al. Immunological status of isolated lymphoid follicles after intestinal transplantation. Am. J. Transplant. 14, 2148–2158 (2014).
Scott, C. L., Wright, P. B., Milling, S. W. F. & Mowat, A. M. Isolation and identification of conventional dendritic cell subsets from the intestine of mice and men. Methods Mol. Biol. 1423, 101–118 (2016).
Dunne, M. R. Flow cytometric analysis of human small intestinal lymphoid cells. WebMD 1326, 61–66 (2016).
Sitohy, B., Hammarström, S., Danielsson, Å. & Hammarström, M. L. Basal lymphoid aggregates in ulcerative colitis colon: a site for regulatory T cell action. Clin. Exp. Immunol. 151, 326–333 (2008).
Gullberg, E. & Söderholm, J. D. Peyer’s patches and M cells as potential sites of the inflammatory onset in Crohn’s disease. Ann. NY Acad. Sci. 1072, 218–232 (2006).
Liebler-Tenoria, E. M. & Pabst, R. MALT structure and function in farm animals. Vet. Res. 37, 257–280 (2006).
Tang, S. & Wu, R. Ilececum: a comprehensive review. Can. J. Gastroenterol. Hepatol. 2019, 1–21 (2019).
Matsui, A. et al. Real-time, near-infrared, fluorescence-guided identification of the ureters using methylene blue. Surgery 148, 78–86 (2010).
Singhal, G. S. & Rabinowitch, E. Changes in the absorption spectrum of methylene blue with pH. J. Phys. Chem. 71, 3347–3349 (1967).
Schreurs, R. R. C. E. et al. Quantitative comparison of human intestinal mononuclear leukocyte isolation techniques for flow cytometric analyses. J. Immunol. Methods 445, 45–52 (2017).
Cornes, J. S. Number, size, and distribution of Peyer’s patches in the human small intestine: Part I The development of Peyer’s patches. Gut 6, 225–229 (1965).
Cornes, J. S. Peyer’s patches in the human gut. J. R. Soc. Med 58, 716 (1965).
Van Kruiningen, H. J., West, A. B., Freda, B. J. & Holmes, K. A. Distribution of Peyer’s patches in the distal ileum. Inflamm. Bowel Dis. 8, 180–185 (2002).
Hellman, T. Untersuchung über die Menge des Lymphoide Gewebes, besonder des Darmes beim Menschen mittels einer quantitativen Bestimmungsmethode. Zeitschrift für die gesamte Anat 2, 191–219 (1922).
Davies, J. et al. Methylene blue but not indigo carmine causes DNA damage to colonocytes in vitro and in vivo at concentrations used in clinical chromoendoscopy. Gut 56, 155–156 (2007).
Hiraku, Y., Goto, H., Kohno, M., Kawanishi, S. & Murata, M. Metal-mediated oxidative DNA damage induced by methylene blue. Biochim. Biophys. Acta 1840, 2776–2782 (2014).
Goodyear, A. W., Kumar, A., Dow, S. & Ryan, E. P. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J. Immunol. Methods 405, 97–108 (2014).
Pimenta, E. M. & Barnes, B. J. Role of tertiary lymphoid structures (TLS) in anti-tumor immunity: Potential tumor-induced cytokines/chemokines that regulate TLS formation in epithelial-derived cancers. Cancers (Basel) 6, 969–997 (2014).
Colbeck, E. J., Ager, A., Gallimore, A. & Jones, G. W. Tertiary lymphoid structures in cancer: drivers of antitumor immunity, immunosuppression, or bystander sentinels in disease? Front. Immunol. 8, 1–18 (2017).
McNamee, E. N. & Rivera-Nieves, J. Ectopic tertiary lymphoid tissue in inflammatory bowel disease: protective or provocateur? Front. Immunol. 7, 308 (2016).
Maoz, A., Dennis, M. & Greenson, J. K. The Crohn’s-like lymphoid reaction to colorectal cancer-tertiary lymphoid structures with immunologic and potentially therapeutic relevance in colorectal cancer. Front. Immunol. 10, 1–9 (2019).
Acknowledgements
We thank all patients and collaborating staff at Herlev hospital and, in particular, the Gastroenterology Team (Department of Pathology) for help in providing tissue samples. We thank A. Mowat (Glasgow University) for valuable feedback during preparation of the manuscript and J. Vandamme for video editing assistance. This work was supported by grants from the Lundbeck Foundation (R155-2014-4184), Denmark, the Gut Cell Atlas, an initiative funded by the Leona M. and Harry B. Helmsley Charitable Trust, US, the Swedish Research Council (2017-02072) and the Swedish Cancerfonden (18 0598) to W.W.A.
Author information
Authors and Affiliations
Contributions
T.M.F., P.B.J., and W.W.A. designed our previously published study. T.M.F. and P.B.J. developed the protocol and performed experiments. U.M.M. performed experiments and discussed data. H.L.J., L.B.R. and O.H.N. provided patient samples and discussed data. P.B.J., T.M.F., U.M.M. and W.W.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Fenton, T. M. et al. Immunity 17, 557–570.e6 (2020): https://doi.org/10.1016/j.immuni.2020.02.001
Extended data
Extended Data Fig. 1 Experimental details.
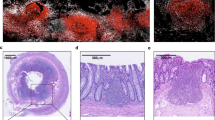
a, Representative image of proximal colonic submucosa after peeling (Step 7) showing SM-ILF (indicated with arrows). Scale bar, 5 mm. b, Immunofluorescence image of a colonic SM-ILF isolated at Step 8. αSMA, alpha smooth muscle actin (green); CD45 (white). Results are from one representative sample of three. c, Diameter (mm) of all colonic SM-ILFs, colonic M-ILFs (cM-ILFs) and ileal M-ILFs (iM-ILFs) detectable in quantified pictures, with bars representing the mean diameter, and each dot representing a single ILF. SM-ILFs were from a total of 68 patients, cM-ILFs from 21 patients, and iM-ILFs from 14 patients. Representative immunofluorescence images of (d-f) peeled LP from proximal colon after Step 7. CD45 (green); DAPI (blue); CD3 (white); CD19 (red). Pictures are representative of three independent samples of which a total 33.9 mm² LP was imaged. Scale bar, 150 µm (a,b) and 1 mm (d–f); triangles indicate the possition of a T cell cluster in (d–f). See supplementary information and source data table for experimental details. GALT, gut-associated lymphoid tissue. LP, lamina propria. M-ILF, mucosal isolated lymphoid follicle. SM-ILF, submucosal isolated follicle.
Extended Data Fig. 2 Innate immune subsets in human GALT and GALT-free LP.
Gating strategy for identifying indicated innate immune subsets in Fig. 3 from GALT-free proximal colon lamina propria (cLP) and colonic submucosal isolated lymphoid follicles (SM-ILF). Green gate, mast cells. Blue gate, granulocytes. Red gate, innate lymphoid cells type-3 (ILC3). Orange gate, dendritic cells (DCs). Purple gate, monocytes/macrophages (Mo/Mac). Histograms showing RORγt staining ILC3 (red) and cells stained with an isotype control (grey). See supplementary information for experimental details. MB, methylene blue.
Extended Data Fig. 3 Biopsy peeling and impact of methylene blue (MB) counterstaining on flow cytometric analysis.
a, Picture taken upon aquisition (Step 1) showing the basal side of two colonic biopsies with visible submucosa (SM), (left) with and (right) without an associated isolated lymphoid follicle (arrow head). b, Picture taken during the peeling process (Step 7) showing the basal side of a colonic biopsy with the SM and associated isolated lymphoid follicle (arrow head) partialy peeled off. c, Histogram showing MB staining intensity in the APC (670/30 nm filter) channel for unstained (grey filled) SM-ILF cells and cells stained with 0.1% (black solid line) or 1.5% MB (dashed line). d, Flow cytomety plot of SM-ILF cells showing non-specific staining of live cells with 0.1% MB in the APC and Alexa flour 700 (730/45 nm filter) channels excited by a red laser (640 nm). Example gate given for MB+ cells. Scale bar, 1 mm. See supplementary information for details.
Extended Data Fig. 4 Identification of Peyer’s patches (PP).
a, Luminal side of PP (outlined) with satellite follicle clusters (arrowhead). (b) and (c) Immunofluorescence images of PP follicles disrupting the muscularis mucosa. αSMA, alpha smooth muscle actin (green); EpCAM, epithelial cell adhesion molecule (red); CD45 (white). d, Complete PP (outlined) with the SM facing up (basal side). e, PP with the SM facing down (luminal side) and associated black spots surrounding individual follicles (arrows). f, PP (outlined) with the SM facing up (basal side) and associated black spots surrounding individual follicles (arrows). (a, d–f) Scale bar, 5 mm. (b,c) Scale bar, 150 µm.
Extended Data Fig. 5 Identification of colonic GALT.
Image of the basal side of proximal colon tissue after submucosal trimming (Step 5) showing examples of visible GALT (arrows). Holes in the tissue produced by cutting are indicated by arrowheads. Scale bar, 1 mm. GALT, gut-associated lymphoid tissue.
Extended Data Fig. 6 Peeling of intestinal samples.
Representative immunofluorescence images of (a) ileum and (b) colonic tissue after Step 2, (c) ileum and (d) colonic lamina propria after Step 7, (e) ileum and (f) colonic submucosa after Step 7, and (g) ileum and (h) colonic M-ILF isolated at Step 13. αSMA, alpha smooth muscle actin (green); EpCAM, epithelial cell adhesion molecule (red); CD45 (white). Pictures are representative of 3 independent samples. Scale bar, 150 µm; (*) indicates the presence of a M-ILF in (a, g and h). See supplementary information for experimental details.
Extended Data Fig. 7 Identification of colonic M-ILF and “craters”.
Image of distal colon after peeling (Step 7) showing examples of M-ILF (arrows) and a “crater” (arrowhead) left behind by a peeled submucosal isolated lymphoid follicle. Scale bar, 1 mm. M-ILF, mucosal isolated lymphoid follicle.
Supplementary information
Supplementary Information
Supplementary Table 1, Supplementary Video Legends 1–8 and Supplementary Methods.
Supplementary Video 1
Turning over full-thickness colon and ileum wall at Step 1, showing the basal and luminal side of each tissue.
Supplementary Video 2
Cutting off fat, serosa and muscularis externa (Step 2) of colon.
Supplementary Video 3
Trimming submucosa (Step 5) of colon.
Supplementary Video 4
Initiation and peeling (Step 7) of colon.
Supplementary Video 5
Initiation and peeling (Step 7) of ileum.
Supplementary Video 6
Excision of methylene blue stained submucosal isolated lymphoid follicles from the peeled submucosa using a tissue punch (Step 12).
Supplementary Video 7
Identification of a submucosal isolated lymphoid follicle from the basal side, and its association with a mucosal invagination on the luminal side, before the initiation of peeling (Step 6).
Supplementary Video 8
Excision of mucosal isolated lymphoid follicles from peeled sigmoid colon lamina propria using a scalpel (Step 12).
Source data
Source Data Fig. 2
Patient-specific viability counts (in %) as well as exact cell-counts of given cell types from given tissues.
Source Data Fig. 3
Patient-specific proportions (% of total CD45+ cells) of given cell types from given tissues.
Source Data Fig. 4
Patient-specific mean diameter and mean concentration of submucosal isolated lymphoid follicles.
Source Data Extended Data Fig. 1
Patient-specific follicle count and size.
Rights and permissions
About this article
Cite this article
Jørgensen, P.B., Fenton, T.M., Mörbe, U.M. et al. Identification, isolation and analysis of human gut-associated lymphoid tissues. Nat Protoc 16, 2051–2067 (2021). https://doi.org/10.1038/s41596-020-00482-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00482-1
This article is cited by
-
Optical imaging of the small intestine immune compartment across scales
Communications Biology (2023)
-
Human intestinal B cells in inflammatory diseases
Nature Reviews Gastroenterology & Hepatology (2023)
-
Characterization of the mucosal microbiota in patients with nodular lymphoid hyperplasia with concurrent irritable bowel syndrome compared to healthy controls
Molecular Biology Reports (2023)
-
Intestinal fibroblastic reticular cell niches control innate lymphoid cell homeostasis and function
Nature Communications (2022)
-
Human gut-associated lymphoid tissues (GALT); diversity, structure, and function
Mucosal Immunology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.