
Role of CYP3A4 in kinase inhibitor metabolism and assessment of CYP3A4 activity
Introduction
Tyrosine kinase inhibitors (TKIs) are small molecules which elicit their effects on tumor (aberrant) cells by entering these and then inhibiting tyrosine kinase (enzymatic) functions. “The small molecules often inhibit multiple enzymatic sites, have a broad spectrum of target kinases, and tend to be substrates of hepatic CYPs with a terminal elimination half-life of 12–24 hours, and thus require daily oral administration” (1). This sentence from Chapter 62 of the “Goodman & Gilman’s” indirectly indicates that there might be some issues concerning the right drug exposure. For most drugs, exposure [area under the plasma concentration time curve (AUC)] is related to a drug receptor interaction which in turn elicits the pharmacological effects influencing the clinical outcome. The most important pharmacokinetic parameter determining drug exposure is drug clearance (drug elimination), because a given drug dose divided by the drug’s clearance results in drug exposure (AUC). The higher the clearance, the lower the exposure. Any interindividual variability in drug clearance therefore results in variable drug exposure and this consequently modulates the pharmacological response.
CYP3A elimination
Most of today’s available drugs are eliminated by members of the CYP3A isozyme subfamily (2). Between subject variability in drug clearance and hence exposure can be quite large and even more variable, if drug-drug interactions interfere with a drug’s clearance. CYP3A activity can be modulated by co-medication over a 400-fold range either by inducing isozyme expression or inhibiting expressed enzymes (3). Therefore, dose adaptation seems to be the logical consequence to ensure optimal drug exposure and thereby drug efficacy in each patient. For some drugs with narrow therapeutic index a therapeutic drug monitoring (TDM) to optimise drug efficacy or reduce drug toxicity is already established. Another approach, mainly used by researchers (4), is to assess the activity of CYP-enzymes with endogenous substrates or probe drugs like midazolam or omeprazole. However, no routine testing of CYP3A activity has been established until today. In drug development all regulatory agencies recommend the thorough investigation of a compound’s potential to be a victim or a perpetrator of CYP (cytochrome P450) enzyme or transporter mediated drug-drug interactions (5,6). However, once a new drug is available on the market, almost no measures are taken to individualise dosing. Instead, some combinations are contraindicated, but for other combinations where a dose adaptation could be used to reduce toxicity risk or enhance efficacy, a simple advice of “Exercise caution with concomitant use of moderate CYP3A inhibitors” is given.
TKIs
Just recently an excellent and elaborate review on the pharmacokinetics of the TKIs has been published where the major factors responsible for the large interindividual variability in exposure of TKIs are discussed (7). Small molecules like TKIs are administered orally which is a huge advantage for patients as they can take the drug on a regular basis at home. Oral administration, however, can be associated with the risk of high first-pass metabolism resulting in much higher doses to be administered in order to ensure the required systemic exposure. This high first-pass metabolism is mainly caused by CYP3A. To highlight the possible consequences, a theoretical example might explain the underlying problems. A drug is administered orally and its absorption is assumed to be complete (100%). The fraction of the dose, which reaches the systemic circulation as parent drug is the bioavailability (F=20%). Hence, the first-pass clearance eliminates 80% of the parent drug, which in this example is assumed to be mediated by CYP3A. Therefore, the individual bioavailability of this drug is inversely proportional to the CYP3A activity. It is therefore possible that bioavailability ranges from 5% to 50% between individuals, resulting in a 10-fold variability of exposure when the same dose is administered. This variability can be even higher if drug interactions occur, inducing or inhibiting CYP3A mediated metabolism. This example clearly shows that we need to know the contribution of CYP3A to the overall clearance of the TKIs. An attempt has already been made (8) and by looking at Table 2, for all but one of the 15 TKIs listed a major contribution of CYP3A was declared, suggesting a strong dependency of drug clearance from CYP3A. However, further clearance mechanisms are not listed. In order to evaluate the importance and relevance of CYP3A for the clearance of the TKIs and hence their drug exposure to patients, all available European public assessment reports (EPAR) (9-20) of 31 TKIs (21-39) were evaluated regarding the clinical pharmacology data with special focus on clearance (6).
Mass balance of TKIs
As a first measure to evaluate clearance, the substances’ route of elimination was assessed. Mass balance studies using radioactive labelled material have been carried out for most of the TKIs and sampling of faeces and urine was performed. To evaluate the total drug clearance in relation to the CYP3A contribution, the first important parameter to know is the fraction of the dose absorbed (fabs). Either this was listed in the EPARs, or it was calculated by adding the amount of drug excreted in urine (which must have been absorbed) to the proportion of metabolites analysed in faeces. However, when parent drug was detected in faeces, it could either be an unabsorbed fraction of the administered dose or an unchanged excreted fraction of the drug via bile into faeces after absorption. Consequently, there is some uncertainty about the faecal excretion of parent drug after oral administration. In fact, the calculated and listed fabs represent the minimal absorbed fraction after oral administration. Of the 31 TKIs, only half are absorbed by at least 50% of the orally administered dose (Figure 1). For dabrafenib, nintedanib, sunitinib, trametinib, and vandetanib, the EPARs do not provide any detailed data on the amounts of parent drug and metabolites excreted in faeces. Secondly, the absolute bioavailability is another important parameter, which has been investigated for 21 of 31 TKIs (11 of them showing less than 50% absolute bioavailability). For substances, which are highly absorbed, but show a low absolute bioavailability, a substantial first-pass metabolism can be anticipated. On the other hand, TKIs with a high bioavailability (>70%, dabrafinib, imatinib, regorafenib, ruxolitinib, and trametinib) are not expected to have a substantial contribution of CYP3A to their elimination in the first place, since less first-pass metabolism is observed. Apart from CYP3A being involved in first-pass metabolism, it also contributes to intrinsic clearance. If other pathways do not play an important role in drug elimination, even drugs with a high bioavailability can undergo CYP3A metabolism for drug elimination. All known bioavailability data are shown in Figure 2 in relation to fabs. From the fraction absorbed and the absolute bioavailability data there is no prediction about the clearance mechanisms involved possible. However, if the drug is highly absorbed, but has a low bioavailability due to a high first-pass metabolism, there is some likelihood that CYP3A is involved. The most likely candidate TKIs would be bosutinib and dasatinib (Figure 2) with fabs of at least 70% (dasatinib) and 55% (bosutinib) and a bioavailability of 34% (both).

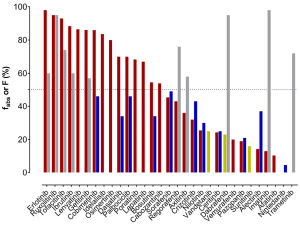
TKIs prone to CYP3A metabolism
As an example, a closer look at bosutinib and dasatinib is taken. Extensive in vitro studies have been carried out with bosutinib (15). CYP3A4 is the predominant enzyme capable of bosutinib metabolism whereas CYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, or 3A5 did not metabolise bosutinib. There is quite a poor description of the metabolism of dasatinib in the EPAR (21). Based on in vitro data, it was claimed that CYP3A4 appears to play a major role in dasatinib metabolism. However, dasatinib is also able to inhibit the activities of CYP2C8 (Ki=3.6 µM) and CYP3A4 (Ki=1.9 µM), the latter one being time dependent. At concentration 10-fold higher than Cmax dasatinib did not induce CYP3A4, 1A2, 2B6, 2C9, and 3A4 in primary cultures of human hepatocytes. For both TKIs data are inconclusive regarding their clearance processes.
Drug-drug interaction studies
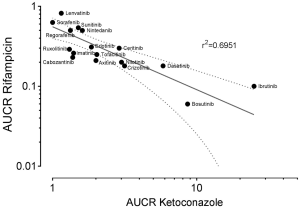
To gain further inside into the role of CYP3A in the metabolism of TKIs, results of drug-drug interaction studies can be used. These studies are mostly part of the drug development programme and the FDA clearly states (5): “The objective of drug-drug interaction studies is to determine whether potential interactions between the investigational drug and other drugs exist and, if so, whether the potential for such interactions indicates the need for dosage adjustments, additional therapeutic monitoring, a contraindication to concomitant use, or other measures to mitigate risk.” In addition, the identification of the major routes of elimination, the relative contribution of enzymes and drug transporters to drug disposition should be carried out. If all in vitro studies have been performed to assess whether a drug is a substrate, inhibitor, or inducer of metabolising enzymes, studies with strong inhibitors and inducers should be carried out to provide a sensitive assessment of the interaction potential. Therefore, if the contribution of CYP3A to the overall elimination of a drug accounts for ≥25% of the clearance, drug interaction studies with a strong CYP3A inhibitor (i.e., ketoconazole) and strong CYP3A inducer (i.e., rifampicin) must be carried out. Most of the TKIs have been investigated using ketoconazole and rifampicin. As an index for the degree of inhibition the ratio of AUC with and without perpetrator (AUCR) is used. AUCR data from 17 TKIs with both ketoconazole and rifampicin are presented in Figure 3. The highest AUCR during ketoconazole was 25 for ibrutinib, the strongest induction by rifampicin was observed for bosutinib (AUCR =0.06). To evaluate a possible relationship between AUCR ketoconazole and AUCR rifampicin a nonlinear regression analysis was performed with the log transformed data of the 17 TKIs available. There was a significant linear regression between both AUCRs (r2=0.6951) (Figure 4), which is not surprising since ketoconazole is a strong inhibitor of CYP3A and rifampicin a strong inducer of CYP3A although other effects might be present additionally. Therefore, if CYP3A is a major contributor of a drugs clearance a high AUCR for ketoconazole and a low AUCR for rifampicin is expected. Of course this relationship can be weakened by other mechanisms. Besides being a strong inhibitor (AUCR ≥5) of CYP3A, ketoconazole is a weak inhibitor (AUCR ≥1.25 but <2) of CYP2C19, CYP2C8, and an inhibitor of the drug transporter P-gp (5). Rifampicin on the other hand is a strong inducer (AUCR ≤0.2) of CYP3A, a moderate inducer (0.2< AUCR ≤0.5) of CYP2B6, CYP2C8, CYP2C9, CYP2C19. It induces the drug transporter P-gp, but also inhibits OAPT1B1 and OATP1B3 (5).
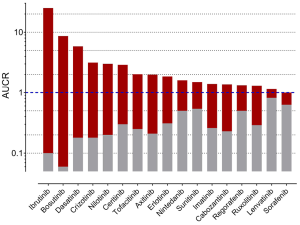
TKIs less prone to CYP3A metabolism
Concerning CYP3A involvement in TKI metabolism, there is clearly an overestimation in published literature, which might have been adopted by some clinicians. Sunitinib might serve as an example. Data extracted from the sunitinib EPAR (35) show that 16% of the dose is excreted via urine and 61% via faeces. The study referred to in the EPAR document reveals that 77% is excreted within 3 weeks after sunitinib administration; however that major portion was excreted during the first 7 days (40). Faecal excretion accounts for 48.2% with 25% of the sunitinib dose being the active metabolite SU012662, followed by the parent drug 13.6%. In urine, similar amounts of sunitinib and SU012662 (6.4% and 6.9%) are found. Improved survival and increased toxicity have been associated with increased exposure to sunitinib (41,42). Depending on the indication patients usually receive a fixed dose of either 37.5 or 50 mg sunitinib once daily. An average sunitinib clearance of about 600 mL/min has been reported by various sources (35,43), dose adjusted trough concentration varied 8-fold for sunitinib and 18-fold for the active metabolite SU012662 (43). “Sunitinib is metabolised primarily by the cytochrome P450 enzyme, CYP3A4, to produce its primary active metabolite, which is further metabolised by CYP3A4” which is a quotation of the prescribing information of Sutent (44). This has led to several studies evaluating the value of CYP3A activity or the influence of pharmacogenetics on sunitinib exposure. A cohort of solid tumor patients (n=52) starting with sunitinib was phenotyped before and during the treatment course for CYP3A and ABCB1 (P-Glycoprotein) using midazolam and 99mTc-MIBI (methoxy-isobutyl-isonitrile) (43). Although there was a correlation between midazolam metabolic ratio and sunitinib pharmacokinetics, it was not sufficient to be useful for a clinical dosing strategy of sunitinib. This is in contrast to an earlier publication, where the authors concluded from a small sample size of 13 patients that midazolam and sunitinib exposure are highly correlated and therefore a large proportion of the observed interpatient variability of sunitinib exposure could be explained by CYP3A variability (45). However, looking at the data provided in the figures, variability of sunitinib (plus its metabolite) was only 2-fold and midazolam variability was 2.5-fold (45). Analysing the drug interaction data available for sunitinib, it must be concluded that sunitinib clearance mechanisms are not yet well understood. The strong CYP3A inhibitor ketoconazole results in an AUCR of 1.51 (35), grapefruit juice as a selective CYP3A inhibitor in the GI-tract showed an AUCR of 1.11 (46), induction of CYP3A by rifampicin caused an AUCR of 0.54 (35). In terms of clearance, ketoconazole will reduce sunitinib clearance from 600 to 400 mL/min and rifampicin will increase clearance to 1,200 mL/min. Knowing that ketoconazole is able to reduce at least 80% of CYP3A activity (47), there is still sufficient clearance available to eliminate sunitinib probably mediated by other still unknown mechanisms.
Future perspectives
For most of the TKIs a fixed dose is used for every patient. Almost no dose recommendations or rules for dose adaptations are given. With respect to drug-drug interactions, TKIs are not listed as drugs or drug classes being prone to life-threatening drug-drug interactions (48). We need to develop indices that will help us to decide when and how to modify the recommended dosing scheme of TKIs. TDM can of course be very helpful in adjusting the dose to achieve the required concentrations. There have been methods developed, where numerous TKIs can be quantified with the same analytical method, thereby facilitating an easy and reliable way to ensure timely drug quantifications (49). Apparently, only a few centres have established TDM for TKI dose optimisation (49). Another possibility might be the use of phenotyping to assess the activity of the principle enzyme(s) involved in metabolism. In order to use phenotyping, it is a prerequisite to have information on the fraction absorbed, the clearance mechanisms involved in elimination, and their relative contribution to the overall clearance. It should be an absolute requirement for drug companies to provide these data to the regulatory agencies. With this knowledge, it will be possible in future to use a probe drug cocktail to assess the individual enzyme and transporter activities at any time necessary. If microdoses are used, the patient is not burdened with drugs and would not suffer from side effects of the probe drugs. This has already been established for midazolam as a probe drug for CYP3A activity (4,50,51); even with a limited sampling strategy involved (52).
Lesson to be learned
If drugs show a relationship between plasma exposure and drug efficacy and/or toxicity clearance pathways must be examined to evaluate the relative contribution of CYP3A to the overall clearance. If CYP3A accounts for less than 25% of total clearance, there is a high probability that no dose adaptation due to CYP3A mediated drug interactions will be necessary. If CYP3A contributes to 50% or more to overall clearance, phenotyping might be applicable in future to adjust the dose to the individual activity and be then validated by TDM.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Michael Sorich and Andrew Rowland) for the series “Precision dosing of targeted anticancer drugs” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.10). The series “Precision dosing of targeted anticancer drugs” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chabner B, Barnes J, Neal J, et al. Targeted therapies: tyrosine kinase inhibitors, monoclonal antibodies, and Cytokines. In: Brunton LL, Chabner BA, Knollmann BC, editors. Goodman & Gilman's The Pharmacological Basis of Therapeutics, Twelfth Edition. NY: McGraw-Hill Education, 2011;1731-69.
- Zhou SF. Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab 2008;9:310-22. [Crossref] [PubMed]
- Backman JT, Kivisto KT, Olkkola KT, et al. The area under the plasma concentration-time curve for oral midazolam is 400-fold larger during treatment with itraconazole than with rifampicin. Eur J Clin Pharmacol 1998;54:53-8. [Crossref] [PubMed]
- Hohmann N, Haefeli WE, Mikus G. CYP3A activity: towards dose adaptation to the individual. Expert Opin Drug Metab Toxicol 2016;12:479-97. [Crossref] [PubMed]
- US-Food and Drug Administration. Guidance for industry: drug interaction studies—study design, data analysis, implications for dosing, and labeling recommendations (draft guidance). 2012. Available online: https://www.fda.gov/downloads/drugs/guidances/ucm292362.pdf
- European Medicines Agency. Guideline on the investigation of drug interactions: final (CPMP/EWP/560/95/Rev. 1 corr. 2). 2012. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2012/07/WC500129606.pdf
- Rowland A, van Dyk M, Mangoni AA, et al. Kinase inhibitor pharmacokinetics: comprehensive summary and roadmap for addressing inter-individual variability in exposure. Expert Opin Drug Metab Toxicol 2017;13:31-49. [Crossref] [PubMed]
- van Leeuwen RW, van Gelder T, Mathijssen RH, et al. Drug-drug interactions with tyrosine-kinase inhibitors: a clinical perspective. Lancet Oncol 2014;15:e315-26. [Crossref] [PubMed]
- European Medicines Agency. Glivec: EPAR - Scientific Discussion. 2004. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000406/WC500022203.pdf
- European Medicines Agency. EMA/CHMP/726072/2014 - CHMP assessment report Vargatef. 2014. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002569/WC500179972.pdf
- European Medicines Agency. EMEA/302222/2008 - ASSESSMENT REPORT FOR TYVERB. 2008. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/000795/WC500044960.pdf
- European Medicines Agency. EMA/491185/2013 - CHMP assessment report Giotrif. 2013. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002280/WC500152394.pdf
- European Medicines Agency. EMA/10821/2017 - CHMP assessment report Alecensa. 2016. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/004164/WC500225709.pdf
- European Medicines Agency. EMA/CHMP/453325/2012 - CHMP assessment report Inlyta. 2012. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002406/WC500132190.pdf
- European Medicines Agency. EMA/70979/2013 - CHMP assessment report Bosulif. 2013. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002373/WC500141745.pdf
- European Medicines Agency. EMA/664123/2016 - CHMP assessment report CABOMETYX. 2016. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/004163/WC500214070.pdf
- European Medicines Agency. EMA/170114/2015 - CHMP assessment report Zykadia. 2015. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003819/WC500187506.pdf
- European Medicines Agency. EMA/685908/2015 - CHMP assessment report Cotellic. 2015. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003960/WC500198565.pdf
- European Medicines Agency. EMA/CHMP/497137/2012 - CHMP assessment report XALKORI. 2012. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002489/WC500134761.pdf
- European Medicines Agency. EMA/CHMP/242419/2013/corr 1 - CHMP assessment report Tafinlar. 2013. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002604/WC500149673.pdf
- European Medicines Agency. Sprycel: EPAR - Scientific Discussion. 2006. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000709/WC500056995.pdf
- European Medicines Agency. Tarceva: EPAR - Scientific Discussion. 2005. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000618/WC500033991.pdf
- European Medicines Agency. EMEA/CHMP/563746/2008 - CHMP assessment report Iressa. 2009. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/001016/WC500036361.pdf
- European Medicines Agency. EMA/CHMP/645137/2014 - CHMP assessment report Imbruvica. 2014. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003791/WC500177777.pdf
- European Medicines Agency. EMEA/CHMP/324336/2014 - CHMP assessment report Zydelig. 2014. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003843/WC500175379.pdf
- European Medicines Agency. EMA/250082/2015 - CHMP assessment report Lenvima. 2015. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003727/WC500188676.pdf
- European Medicines Agency. Tasigna: EPAR - Scientific Discussion. 2007. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000798/WC500034398.pdf
- European Medicines Agency. EMA/CHMP/15445/20165 - CHMP assessment report TAGRISSO. 2015. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/004124/WC500202024.pdf
- European Medicines Agency. EMA/652627/2016 - CHMP assessment report IBRANCE. 2016. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003853/WC500217198.pdf
- European Medicines Agency. EMA/CHMP/248579/2010 - CHMP assessment report Votrient. 2010. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/001141/WC500094275.pdf
- European Medicines Agency. EMA/CHMP/220290/2013 - CHMP assessment report Iclusig. 2013. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002695/WC500145648.pdf
- European Medicines Agency. EMA/CHMP/403683/2013 - CHMP assessment report Stivarga. 2013. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002573/WC500149166.pdf
- European Medicines Agency. EMA/465846/2012 - CHMP assessment report Jakavi. 2012. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002464/WC500133226.pdf
- European Medicines Agency. Nexavar: EPAR - Scientific Discussion. 2007. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000690/WC500027707.pdf
- European Medicines Agency. Sutent: EPAR - Scientific Discussion. 2007. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000687/WC500057733.pdf
- European Medicines Agency. EMA/CHMP/853224/2016 - CHMP assessment report Xeljanz. 2017. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/004214/WC500224913.pdf
- European Medicines Agency. EMA/CHMP/258608/2014 - CHMP assessment report Mekinist. 2014. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002643/WC500169708.pdf
- European Medicines Agency. EMA/128076/2012 - CHMP assessment report Caprelsa. 2012. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002315/WC500123603.pdf
- European Medicines Agency. EMA/200986/2012 - CHMP assessment report Zelboraf. 2012. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002409/WC500124400.pdf
- Speed B, Bu HZ, Pool WF, et al. Pharmacokinetics, distribution, and metabolism of [14C]sunitinib in rats, monkeys, and humans. Drug Metab Dispos 2012;40:539-55. [Crossref] [PubMed]
- Houk BE, Bello CL, Poland B, et al. Relationship between exposure to sunitinib and efficacy and tolerability endpoints in patients with cancer: results of a pharmacokinetic/pharmacodynamic meta-analysis. Cancer Chemother Pharmacol 2010;66:357-71. [Crossref] [PubMed]
- Houk BE, Bello CL, Kang D, et al. A population pharmacokinetic meta-analysis of sunitinib malate (SU11248) and its primary metabolite (SU12662) in healthy volunteers and oncology patients. Clin Cancer Res 2009;15:2497-506. [Crossref] [PubMed]
- Kloth JS, Binkhorst L, de Wit AS, et al. Relationship Between Sunitinib Pharmacokinetics and Administration Time: Preclinical and Clinical Evidence. Clin Pharmacokinet 2015;54:851-8. [Crossref] [PubMed]
- US-Food and Drug Administration. SUTENT: Prescribing information. 2015. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/021938s028s029lbl.pdf
- de Wit D, Gelderblom H, Sparreboom A, et al. Midazolam as a phenotyping probe to predict sunitinib exposure in patients with cancer. Cancer Chemother Pharmacol 2014;73:87-96. [Crossref] [PubMed]
- van Erp NP, Baker SD, Zandvliet AS, et al. Marginal increase of sunitinib exposure by grapefruit juice. Cancer Chemother Pharmacol 2011;67:695-703. [Crossref] [PubMed]
- Fuchs I, Hafner-Blumenstiel V, Markert C, et al. Effect of the CYP3A inhibitor ketoconazole on the PXR-mediated induction of CYP3A activity. Eur J Clin Pharmacol 2013;69:507-13. [Crossref] [PubMed]
- Day RO, Snowden L, McLachlan AJ. Life-threatening drug interactions: what the physician needs to know. Intern Med J 2017;47:501-12. [Crossref] [PubMed]
- van Dyk M, Miners JO, Kichenadasse G, et al. A novel approach for the simultaneous quantification of 18 small molecule kinase inhibitors in human plasma: a platform for optimised KI dosing. J Chromatogr B Analyt Technol Biomed Life Sci 2016;1033-1034:17-26. [Crossref] [PubMed]
- Halama B, Hohmann N, Burhenne J, et al. A nanogram dose of the CYP3A probe substrate midazolam to evaluate drug interactions. Clin Pharmacol Ther 2013;93:564-71. [Crossref] [PubMed]
- Hohmann N, Halama B, Siller N, et al. Response to "can CYP3A activity be evaluated for drug interaction using a nanogram dose of probe drug?": evaluation of CYP3A activity with microdoses of midazolam. Clin Pharmacol Ther 2014;95:490-1. [Crossref] [PubMed]
- Katzenmaier S, Markert C, Riedel KD, et al. Determining the time course of CYP3A inhibition by potent reversible and irreversible CYP3A inhibitors using a limited sampling strategy. Clin Pharmacol Ther 2011;90:666-73. [Crossref] [PubMed]


