Related Factors to Streptococcus pneumoniae Invasive Infection and Clinical Manifestations: The Potential Role of Nasopharyngeal Microbiome
- 1Infectious Diseases Unit, Hospital Universitari Mútua Terrassa, Terrassa, Spain
- 2Department of Medicine, Universitat Internacional de Catalunya, Barcelona, Spain
- 3Institut de Recerca Sant Joan de Déu, Hospital Sant Joan de Déu, Barcelona, Spain
- 4Center for Epidemiology and Public Health, CIBERESP, Instituto de Salud Carlos III, Madrid, Spain
- 5Internal Medicine Department, Clínica Rotger, Palma de Mallorca, Spain
Infections of the lower respiratory tract, such as pneumonia, are one of the leading causes of death worldwide. Streptococcus pneumoniae might colonize the upper respiratory tract and is the main aetiological agent of community-acquired pneumonia (CAP). In the last decades, several factors related to the host, the microorganism and the antibiotic therapy have been investigated to identify risk factors associated with the development of invasive pneumococcal disease (IPD). Nevertheless, these factors themselves do not explain the risk of developing disease or its severity. Recently, some studies have focused on the importance of nasopharyngeal (NP) microbiome and its relation to respiratory health. This review presents existing evidence of the potential role of NP microbiome in the development of IPD.
Introduction
Lower respiratory infections, considering also pneumonia, represent the third cause of death worldwide, with a median of 4.2 million deaths (7.1% of total deaths) per year (1). Combination of pneumonia and Influenza ranked 8th among leading causes of mortality in developed countries. Also, they are the main cause of death due to infection (2). Streptococcus pneumoniae (S.pneumoniae) is the most common pathogen implicated in community-acquired pneumonia (CAP) and accounts for two-thirds of the mortality in hospitalized patients (3).
Invasive pneumococcal disease (IPD) refers to an infection confirmed by the isolation of S.pneumoniae from a normally sterile site (pneumonia, parapneumonic empyema, meningitis, bacteraemia/sepsis, peritonitis and arthritis) (4). Its incidence, despite its remarkable reduction since the introduction of pneumococcal conjugated vaccine (PCV), is still 10–20/100,000/year in developed countries and could be even higher in developing countries (5–11).
To date, the risk of developing pneumococcal pneumonia and also IPD has been associated with the interplay between host susceptibility and pathogen virulence.
Many factors have been described regarding host susceptibility for the development and prognosis of pneumococcal pneumonia, as age, comorbidities and previous vaccination status (12–15). Severity and mortality in pneumococcal pneumonia are also related to strain characteristics, coinfection and antibiotic treatment (16, 17).
However, these elements do not fully explain by themselves the potential risk for infection and its severity. Despite recent advances in this field, it is still not known why some individuals present asymptomatic NP colonization; others develop localized infection and others, IPD.
The aim of this review was to present the current evidence for the role of different factors –including the potential role of NP microbiome- involved in the dissemination of S.pneumoniae from the nasopharynx to other human body sites and its subsequent clinical manifestations.
Methods
A literature search of PubMed and WOS databases conducted between January 2000 and February 2021 using the parameters [microbiome (MeSH Terms)] OR [infection, streptococcus pneumoniae (MeSH Terms)] OR [pneumonia (all fields)] generated >180,000 abstracts.
Titles and abstracts were assessed, as a rule of thumb, and clinically relevant articles were reviewed. References of the selected articles were also reviewed and those which could fit the topic were read and evaluated as well.
Host Characteristics
Baseline Characteristics and Clinical Features
S.pneumoniae is a large contributor to mortality worldwide (12, 13, 18). It is the most common documented etiology of severe bacterial respiratory tract infection in children, but also in the adulthood. The spectrum of disease ranges from colonization (especially among children under 5 years of age) to mucosal disease (otitis media, sinusitis, pneumonia), and to invasive infections (bacteremia, meningitis, endocarditis, others) (18).
Increasing age, male gender, toxic habits (as smoking and alcohol abuse), liver or renal disease, solid organ tumors, immunosuppression (HIV infection, asplenia) and higher Charlson Comorbidity Index have been identified as independent risk factors for high mortality among patients with community-acquired, bacteraemic pneumococcal pneumonia.
Among all the host factors previously described, age has been identified as the strongest predictor of death, even in patients without significant comorbidity (13, 19, 20).
Vaccination
S.pneumoniae infection is one of the most vaccine-preventable diseases. In the 1980s and 1990s, the 23-valent-polysaccharide vaccine (PPSV23) was the only direct prevention measure available (12). Polysaccharide antigens produce the activation of mature B cells. However, protein-polysaccharide conjugate pneumococcal vaccines (PCV) have a T-cell-immune response and are effective in immunosuppressed patients (21). In 2000 the 7-valent vaccine (PVC7) -against seven serotypes- was licensed for children in the USA. After that, the increment in relevance of non-vaccine serotypes, conducted to the development of new vaccines: PCV10 (PCV7 plus 1, 5 and 7F), and PCV13 (PCV10 plus 3, 6A and 19A) (22). At present, there are two new conjugate vaccines in development (PCV15 and 20vPnC) (23).
Many investigators have focused on the subsequent impact of young children vaccination in the incidence of IPD in other age groups. Although there are some reports of this incidence being increased (24, 25), there is solid evidence of a sustained decrease in IPD incidence in vaccinated children and adults, -including the immunosuppressed population- supporting the use of PVCs (12, 22, 26–29).
Genetic Polymorphisms
The innate immune system represents the first non-specific step in host defense. The recognition of pathogens by the host immune system is a necessary requisite for the initiation of a response (15). A bunch of different receptors are placed on the cell surface of the epithelial barrier and on hematopoietic cells. These receptors recognize diverse pathogen antigens. After these pattern recognition, a very complex net of intracellular signaling pathways is triggered in order to develop a response of the host against the pathogen (30).
In the 1980s, genetics was found to be a major determinant of susceptibility to infectious diseases. Extreme-phenotype studies in patients with recurrent IPD were successful in the identification of factors associated with increased susceptibility (31). Different genetic variants in the proteins involved in the signaling pathways have been identified in the last 20 years as elements that either increase the risk or confer protection against pneumococcal pneumonia. A recent meta-analysis showed that variants in CD14 and MBL2 genes were associated with susceptibility to pneumococcal disease. Several other host genetic polymorphisms have been identified that potentially influence susceptibility and outcome of pneumococcal disease, although most of these findings have not been confirmed in independent studies (32).
Thus, all these findings support the hypothesis that genetic variants may explain, at least partially, the host susceptibility to pneumococcal pneumonia and IPD.
Bacterial and Treatment Factors
The outcome of IPD can be affected by host factors, such as age (the very young and the very old), underlying conditions, low socioeconomic status and quality of life; but also by bacterial factors such as the serotype (16).
Serotype, Invasiveness, and Severity
At least 100 serotypes of S.pneumoniae has been identified, based on antigenic differences in their capsular polysaccharides (18).
Main serotypes found in NP carriage and invasive disease are quite similar worldwide (33, 34). Likewise, the frequency by which a certain serotype causes invasive disease per carriage episode is a stable property along the time. There is an inverse relationship between frequency of carriage and invasiveness: serotypes which are less commonly carried cause more frequently invasive disease whilst the serotypes most prevalent in carriage are less invasive (35). Also, it has been described that serotypes which have less risk of causing invasive disease are associated with more severe disease and mortality, even in healthy individuals (36–38).
According to the studies of Brueggemann et al. serotypes 1, 3, 4, 5, 7F, 8, 9A, 9V, 12F, 14, 18C, and 19A are considered highly-invasive serotypes while the remaining serotypes were considered non-highly-invasive or opportunistic (39).
A meta-analysis of IPD outcome by serotype in 2011 was consistent with these findings: there is a relationship between IPD and pneumococcal serotype. The risk for IPD has a direct relation with the serotype prevalence, and is inversely correlated with invasiveness. Besides, the findings of this study suggested a potential mechanism for the epidemiologic relationships between serotypes. The most prevalent among carriage isolates are the more heavily encapsulated, which rarely cause bacteraemic invasive disease, but can cause more severe disease when they do invade (16).
Impact of Initial Antimicrobial Treatment
The effect of S.pneumoniae resistance to antimicrobials and subsequent discordant antimicrobial therapy (DAT) has been largely investigated with controversial results. While some studies observed that patients who received DAT did not have a higher mortality rate (40), others concluded that discordant antibiotic prescribed at admission was strongly related with higher mortality (41).
More recently, in a study conducted in Spain to assess the relationship between empirical antibiotic treatment and mortality in CAP due to S.pneumoniae, the authors evaluated some clinically relevant situations. Among the subgroup of bacteraemic patients, the choice of empirical antibiotic combination other than monotherapy with β-lactam or macrolide combination of β-lactam and macrolide or levofloxacin alone or in combination was associated with higher mortality (17).
Regarding beta-lactam therapy, pharmacokinetic and pharmacodynamic studies suggest that there is no association between mortality and penicillin MIC ≥2 mg/mL in patients that receive penicillin monotherapy at a dosis of ≥10 MU per day (usually ≥12 MU per day) (42).
Nasopharyngeal Microbiome and Respiratory Disease
The human body is considered to be a super-organism which is composed of more microbial cells than body cells (43). The term microbiome was suggested by Joshua Lederberg and refers to the collective genomes of our endogenous microbes or microbiota. This human microbiota plays essential roles in human body functions, as the maintenance of the integrity of the epithelium (44), the modulation of the immune response (45, 46), and the “colonization resistance” to avoid the invasion of pathobionts, microorganisms which can live as commensal symbionts but, under not yet completely characterized circumstances (related to host genetics, microbial context, etc.) could act as pathogens and cause invasive disease (47–52). Research on this topic has largely focused on the gut microbiota; however, recent studies provide growing evidence of the importance of respiratory ecosystems on human health.
Classically, the lung has been considered sterile. More recently, the use of molecular typing [especially quantitative PCR for 16S ribosomal RNA (rRNA)] has allowed identification of pathobionts in culture-negative respiratory specimens (53, 54). These techniques have permitted the identification of five phyla in the lungs of healthy individuals: Actinobacteria, Proteobacteria, Bacteroidetes and Firmicutes (55), including low levels of oral bacteria, like those of the genera Prevotella and Veionella (56, 57). The composition of the lower respiratory tract (LRT) microbiota seems to depend on the upper respiratory tract (URT) microbiota composition, due to aspiration of oropharyngeal secretions, micro aspiration (i.e., while sleeping) or direct contact by continuous mucosa (48, 58, 59). Some studies report that there are indistinguishable microbiota patterns along the respiratory tract in healthy individuals, with decreasing biomass content from upper to lower respiratory tract (56, 60). One study found high viral and bacterial microbiota concordance between nasopharyngeal and endotracheal samples during the course of childhood LRTIs, suggesting that upper respiratory tract (URT) samples could be a valid proxy for lung microbiota during disease states (61). These findings have now changed the assumption that the lower airways are normally sterile (62).
Lung colonization by microbiota starts after birth and therefore remains unaltered during the whole lifetime (63–65). Gut and lung microbiota are actually connected in the gut-lung immunity axis. The metabolites produced by the gut microbiome (short-chain fatty acids) can reach other organs and have some influence in the respiratory diseases (66, 67). Recently, it has been shown the role of the gut-lung immunity axis in the pneumococcal pneumonia, by whom the integrity of the gut microbiota plays a main role in the host defense (68).
The colonization of the URT is the essential first step for a respiratory infection, either of the URT, LRT or a disseminated infection (33). Between the different species of the microbiota, there can exist positive interactions (as mutualism or commensalism) or negative interactions (antagonism) (48). Thus, some members of the microbiome have potential advantageous effects on ecosystem equilibrium, health and functionality (69). Examples of these members in the URT microbiota are Dolosigranulum spp. and Corynebacterium spp., as they have been related to respiratory health and negatively associated to disease caused by pathobionts, such as S.pneumoniae (48, 70, 71). The impediment for the pathogens to find necessary resources because the available nutrients might be used by a diverse local microbiome is one of the mechanisms described behind colonization resistance (48). Certain microorganisms have clearly been identified as having the ability to exclude pathogens from the NP ecosystem (72). In the case of S.pneumoniae, for instance, the release of free fatty acids -which come from the skin surface tiacylglycerols- by the Corynebacterium species C.accolens, has been described as one of the mechanisms which could inhibit pneumococcal growth (71). Furthermore, colonization resistance may be enhanced by interactions with the host immune system (48).
The URT is considered to be a major reservoir for potential pathogens, including S. pneumoniae, from where it could expand and arrive to the lungs and potentially cause infection (33). Thus, the study of the microbiome in the respiratory tract will shed light in the understanding of susceptibility to pulmonary infections, pneumococcal pneumonia or even IPD.
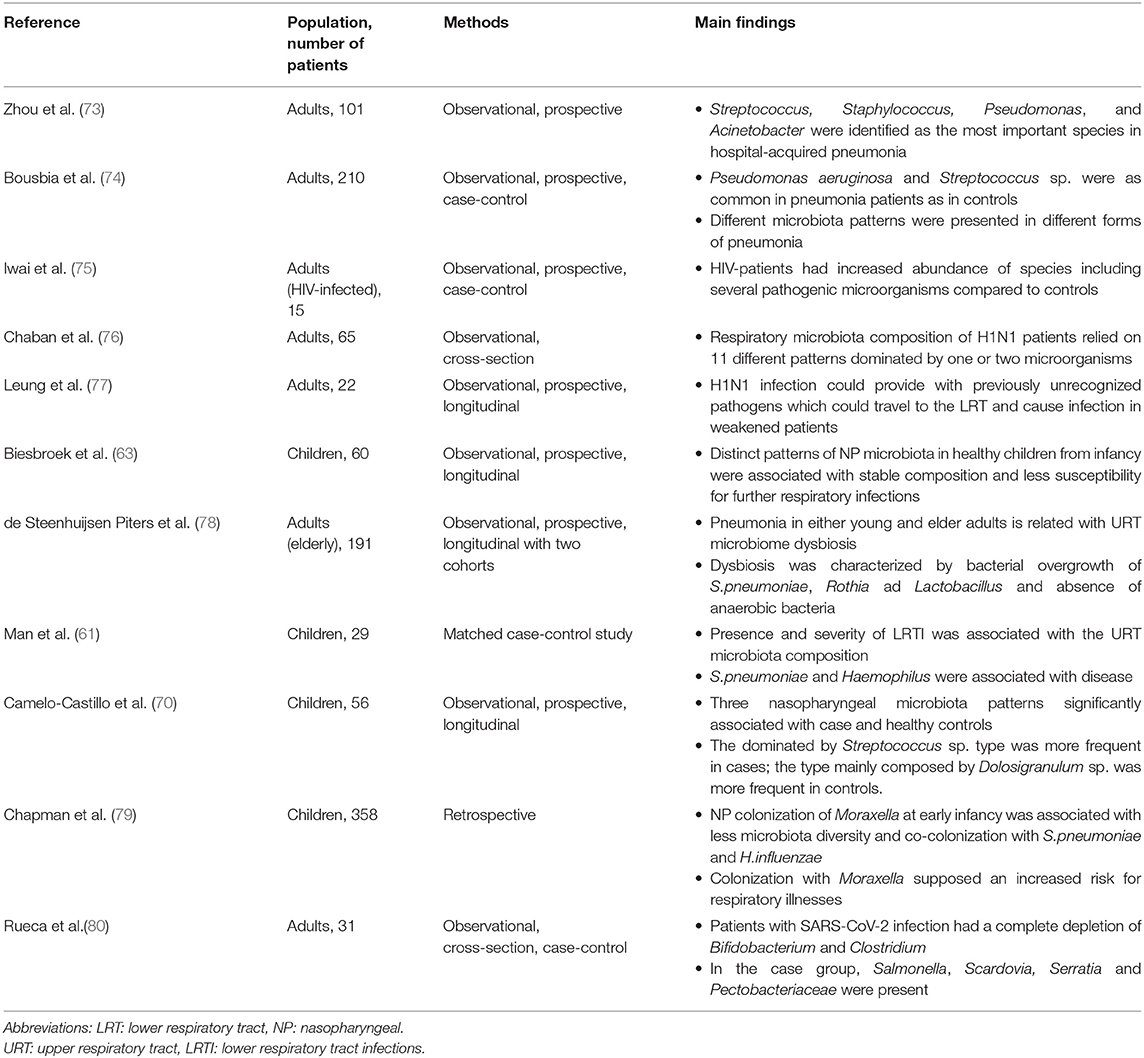
Main advances on this field have been achieved on pediatric population and adult population with chronic respiratory diseases, as chronic obstructive pulmonary disease (COPD). A relation of recent studies addressing the relationship between respiratory microbiome and infection is showed in Table 1. In samples from the respiratory tract of children, it has been described a high inter-individual variability in bacterial composition (81). Despite this variability, the bacterial composition has been proposed to be within discrete categories where some bacterial taxa dominate the community. In infants between 1 and 3 months of age, the hypopharynx was found to have five “pneumotypes” which follow concrete trajectories during the child's development (82). Distinct microbiota profiles have been detected in young children (under 2 years old). Early-life profiles are associated with microbiota stability and change patterns of change during the two first years of life. During the first 2 years, stable microbiota profiles were characterized by an early colonization of Moraxella, and Dolosigranulum combined with Corynebacterium, whereas instability was associated with profiles dominated by Haemophilus and Streptococcus. Moreover, infant feeding and frequency of respiratory infections in the parents were associated with the patterns of change. A history of breastfeeding and a reduced number of consecutive respiratory infections were associated with stable microbiota profiles (63).
Studies of the early URT microbiota in children have connected the early microbiota to the development of disease later in life and to the impact of important drivers. Accelerated microbiota maturation (enriched of Neisseria spp. and (facultative) anaerobes, mainly oral species including Prevotella) is associated with microbiota instability and increasing number of respiratory tract infections (RTIs) over the first year of life. These changed dynamics could be observed within the first month of age and, as stated previously, might be connected to important drivers: a healthy microbiota development and stability might be linked to vaginal delivery and breastfeeding, although this connection is controversial (83, 84).
In 2020 Chapman et al. conducted a large, retrospective study in children in order to address the reasons why some infants are more susceptible to respiratory infections than others (79). In this study, the cohort was divided into two groups: “infections and allergy prone group (IAP)” and “non-infection and allergy prone group (NIAP).” Males and daycare attendance were independently identified as risk factors for IAP. In terms of microbiome characteristics, colonization of NP in an early-age (between 6 and 36 months) with any of the three pathobionts most frequently associated with respiratory infections (S.pneumoniae, H.influenzae and M.catarrhalis) was associated with the IAP group.
More concretely, regarding the potential relationship between pneumococcal infection and microbiome, a case-control study was conducted in Spain to compare the nasopharyngeal microbiota of children with IPD (cases) and healthy children (controls) -representative of a healthy nasopharyngeal niche. In this study, bacterial richness and diversity were significantly higher in the groups of children who developed IPD. Three clusters corresponding to three different nasopharyngeal-types (nasopharyngeal types A, B or C, respectively) were observed. These patterns were significantly associated with the classification of the patients into cases and controls. The most frequently detected pattern in children with IPD (observed in 50.0% of the cases) was nasopharyngeal-type B, mostly represented by the genera Streptococcus (36.9%), Staphylococcus (21.3%), Veillonella (9.8%) along with a diversity of anaerobic genera (Prevotella and Porphyromonas). The other two nasopharyngeal types [type C, composed of Haemophilus (52.1%), Moraxella (31.4%) and Streptococcus (11.4%)] and type A [composed mainly of the genera Dolosigranulum (44.3%), Moraxella (29.3%) and Haemophilus (10.5%)] were detected in 32.1 and 17.9% of children with IPD, respectively. Conversely, the nasopharyngeal-type A was the most frequently related with healthy controls, leading to the hypothesis—and according to previous studies (63)—that Dolosigranulum sp. could confer resistance against pneumococcal infection (70). A clear imbalance was observed with a high frequency of Veillonella and other oral microorganisms which were be relatively infrequent in controls. These results were surprising given that higher bacterial diversity has been associated with health, and lower bacterial diversity has been associated with disease (85).
In adults, a recent study was designed to evaluate the characteristics of the NP microbiota in the pneumococcal acquisition (86). At baseline, samples from the NP of the healthy volunteers enrolled, showed mainly species from four genera: Corynebacterium, Dolosigranulum, Staphylococcus and Streptococcus. From these frequently found bacteria, the NP microbiome could be divided into five different profiles (A-E). Profiles B-E were dominated by Staphylococcus sp., Streptococcus and Corynebacterium sp. or Corynebacterium sp. and Dolosigranulum sp. combined, whilst profile A showed more diversity. The latter profile was more frequently associated with S.pneumoniae carriage (86). In other study, where bronchoalveolar lavage samples were analyzed, two pneumotypes in asymptomatic adults were found: one of them, enriched with supraglottic-characteristic bacteria Prevotella and Veillonella was associated with higher levels of inflammatory markers (57).
A large revision on the role of microbiome in the innate immune response related to chronic lung diseases (CLD) was published in 2020 (87). COPD is a progressive inflammatory disorder characterized by persistent airflow limitation, obstructive bronchiolitis and parenchymal destruction (88) with a high disease burden and related mortality rates. An specific pattern of microbiota has been identified in COPD patients, in comparison with healthy controls (87). Proteobacteria were more frequent in COPD than Bacteroidetes (and Prevotella spp. was specially reduced) (89). COPD mortality is higher during periods of acute exacerbations (AECOPD). During AECOPD, patients show a temporally dynamic sputum microbiome (90). In addition, the sputum microbiome profile at first day of exacerbation is related to 1-year mortality. The absence of Veillonella increases mortality risk by 13-fold whereas the presence of Staphylococcus increases the risk by 7-fold. When these two factors were combined in the same individual, 1-year mortality risk in COPD increased by 85-fold (91). Thus, reduced diversity of microbiome in sputum of AECOPD patients confers a poor prognosis, which is consistent with the previous association of higher bacterial diversity and health (90, 91).
Asthma has largely been studied on its relationship with microbiome. Several studies have identified, on one hand, the respiratory microbiota pattern related to asthma (high abundance of Haemophilus influenzae, Streptococcus pneumoniae, Staphylococcus aureus, and Moraxella catarrhalis, which can potentially play a pathogenic role) (87, 92) and on the other, that these species have a role on the inflammatory response which could determine the outcomes in asthma (93).
Apart from CLD, a limited number of studies have investigated the possible influence of the URT microbiome on the development of LRTIs in adult patients. In general, published studies t include moderate sample sizes without a healthy control group (73, 74), or specific risk groups like HIV (human immunodeficiency virus)-infected patients (75), or patients infected with the pandemic H1N1 influenza virus (76, 77).
In the last year, the spreading of SARS-CoV-2 a novel beta-coronavirus, has provoked a world pandemic. COVID-19, the disease caused by this new virus, ranges from an asymptomatic estate to a severe pneumonia associated to a potentially lethal adult respiratory distress syndrome (SDRA) (94). In the search of answer to the question of why this respiratory virus could affect humans in such a different way, the role of respiratory microbiota has been put on the spot. A very recent review about the role of respiratory microbiota in COVID-19 patients (95) concluded, based on previous evidence, that the dysbiosis in the microbiota in COVID-19 patients would potentially lead to infection or progression of the disease. However, studies on the characteristics of microbiome in COVID-19 patients are still scarce and conducted in a small number of patients to make them suitable for extrapolating conclusions (80, 96).
Although it seems reasonable that the respiratory microbiome composition may play a role in the development of IPD, there is, to our knowledge, one single study that has investigated this association in elderly and young adults (78). In this study by de Steenhuijsen et al. the differences in microbiota profiles between patients with pneumonia and their healthy controls were identified as an independent factor. In the elderly, pneumonia was associated with Rothia, Lactobacillus and Streptococcus (pseudo) pneumoniae whilst healthy adults showed greater diversity and higher richness of especially three different patterns of microbiome (Prevotella melaninogenica, Veillonella and Leptotrichia) in the oropharynx samples (78).
Therapeutic Options in Respiratory Infections
One of the main applications of the knowledge of microbiota composition are the potential therapeutic options. The modification of microbiota profiles to a protective pattern which could lead to a less degree of tissue inflammation, damage and therefore disease, is an attractive approach for this novel research (97).
A previous systematic review evaluated 23 trials in children to evaluate the efficacy of probiotics for prevention and treatment of recurrent respiratory infections (RRI) (98). After meta-analysis and considering the available evidence, probiotics were postulated as a possible alternative therapeutic option for RRI in children. However, the probiotic strain, dosage and administration forms were very heterogeneous among the different studies analyzed.
Human-associated microorganisms are able to produce secondary or specialized metabolites (natural products) which could mediate in the interactions between host and microbes and between microbes themselves (99). These natural products released by non-pathogenic species could be a novel source of antimicrobials, due to their antimicrobial activity against pathogen species (99, 100). More recently, Manti et al. conducted a prospective study in order to prove the a priori protective role of Streptococcus oralis and Streptococcus salivarius using them as a therapeutic option (101). Ninety-one children between 1 and 12 years old were prospectively included in a single-open study, in which a nasal spray composed of Streptococcus oralis 89a and Streptococcus salivarius 24SMBc was administered. After probiotic treatment, clinical improvement was reported, even in children with previous history of atopy or allergies. However, results were only applicable for the initial 3-months, due to a lack of follow-up period in the study protocol.
Phage therapy has arisen as a promising therapeutic tool in the era of antimicrobial resistance (102). Phage lysins, which are encoded by phages, are cell wall hydrolases that selectively act against different peptidoglycan bonds. These proteins can attack especially Gram-positive bacteria by splitting the bacterial cell wall (103). Some lysins have demonstrated activity against S.pneumoniae (104). In a recent research, a new phage lysine, 23TH_48 has been postulated as a potential therapeutic weapon for pneumococcal infections, combined with other lysins or antibiotics. This sinergyc combinations could be used to broaden the spectrum of action and improving their antimicrobial activity (103).
Discussion
Recent advances in molecular typing techniques have identified not only that the lungs are not sterile –as was classically believed- but also that the respiratory tract is colonized by microbial species, which change between healthy individuals and those affected with respiratory diseases (53–57). Microbial populations play an important role in health. Along the human airways, structures above the vocal cords are exposed to high bacterial burden producing contamination of lower airway secretions from the URT (57).
We have reviewed the most important factors known to be associated with pneumococcal disease and we have focused on the available evidence of the role of respiratory microbiome in the development of respiratory infections in children and adults. It has not been established how these previously identified factors might impact in respiratory microbiota development and thereby in susceptibility to LRTIs. Despite the many comorbidities and conditions that have been identified as risk factors for the development of IPD, only the extremes of age (<5 years and >60 years old) have been strongly consistently found to be a major risk for IPD (13, 19, 20). Lung colonization is believed to start early after birth, with different profiles related with different factors, such as infant feeding. Stable patterns were associated with less risk of respiratory infections whilst changing patterns were associated with increased incidence of respiratory infections (63). The presence of these changing microbiota patterns–with Haemophilus and Streptococcus dominant profiles- in children under 2 years of age could explain the higher incidence of IPD in this population. On the other hand, changes on respiratory microbiota through age in the adults were also associated with a higher risk for LRTI. The absence of anaerobic species in the very old –a phenomenon linked to increasing age- could be associated with a high susceptibility for pneumonia at the extreme of life (78).
Vaccination against pneumococcal has changed the epidemiology of the pneumococcal infections and, despite controversial results of previous studies, there has been a decrease in IPD incidence in the vaccinated population, even in the immunosuppressed (12, 22, 26–29). Pneumococcal vaccination has had also an impact in NP microbiota characteristics. In spite of a lack of evidence for a different composition between vaccinated and non-vaccinated children, a higher abundance was identified in patients after PCV 10 vaccine. This complexity could explain that, after vaccination, individuals are less prone to suffer acute respiratory tract infections (105).
Several host genetic polymorphisms which control the pathways of the immune system to combat bacterial infection have been identified as a risk factors for protection or susceptibility to pneumococcal pneumonia (32). Considering that some factors -as, for example, CXCL16- are regulated by microbiota through modulation of the quantity of iNKT cells in the gut and lung, leading to a higher tissue inflammatory response (46), these genetic variations could lead to a different expression of signaling proteins which could be, in turn, modified by the different microbiota patterns conferring more risk or protection against pneumococcal pneumonia.
Studies in recent years have focused on characterization of the respiratory microbiota, and concluded that the LRT microbiota composition comes from the URT (48, 53–59). Due to its role in the regulation of the immune response and inflammation (45, 46), the respiratory microbiome has been associated to the development and exacerbations of chronic lung diseases, as asthma and COPD (87–93). However, the most relevant findings in this review have been the implication of respiratory microbiome in pneumonia. Thus, colonization of the NP with either S.pneumoniae, H.influenzae or M.catarrhalis in children was associated with a tendency for respiratory infections and allergy in the pediatric period. Moreover and related to the scope of this review, in the study conducted by Camelo-Castillo et al. different microbiota patterns on the NP of children, were associated to IPD or asymptomatic colonization in this population (70).
Conversely, in the adult less is known on the impact of microbiome development of LRT infections, including pneumonia and more specifically, pneumococcal pneumonia and IPD. De Sreenhuijsen et al. found -in contrast with the previous studies in children- that anaerobic species were highly represented in old patients with pneumonia, who also had a less diverse and rich oropharyngeal microbiota profile (78).
The identification of microbiota profiles associated to IPD or asymptomatic colonization may be of clinical value as disease biomarkers. According to recent encouraging data about the potential role of probiotics in the treatment and prevention of respiratory infections (mainly in pediatric population) (97, 98, 101), the characterization of beneficial bacteria in adults -preventing or protecting against pneumococcal infection- would allow integrating those microorganisms in a probiotic preparation for the treatment or prophylaxis of pneumococcal infections and IPD.
The main limitation of our study is that we have tried to conduct an unbiased, accurate review of the most relevant literature regarding the evidence on the role of the respiratory microbiota in the development of pneumococcal pneumonia and especially, IPD. However, some relevant publications could have gone unnoticed by our research system and therefore not included in the current literature review.
Finally, considering that the microbial ecosystem of adults is relatively stable in the absence of gross perturbations, the role of microbiome in IPD in children might be likely reproducible in adults.
Data regarding the potential relationship between NP microbiome and the risk of developing IPD in adults, viral coinfection and severity of disease are scarce, and specific research in this area is needed. Although NP microbiome in patients with IPD has not been properly characterized yet, there seem to be discordant results between pediatric and adult populations. New, longitudinal studies, with larger number of participants and a homogeneous system to collect samples should help to elucidate the potential role of the previously observed microbial species in adults and their relationship with increasing or reducing risk for the development of respiratory infections, especially IPD.
Data Availability Statement
The original contributions generated for the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
DH, EC, LB-P, and CM-A contributed to the conception and the design of the review. DH, EC, LB-P, and BD contributed to the literature research and selection. JG contributed to the coordination of the review. BD and EC were the major contributors in writing the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported in part by Ministry of Science and Innovation, Instituto de Salud Carlos III (ISCIII) (http://www.isciii.es), Projects of Research on Health (FIS project number PI19/01875, Principal Investigator: EC and FIS project number PI16/00174, Principal Investigator: CM-A) and Pre-doctoral Contract for training in research into Health (Grant number FI17/00248, Recipient: DH) by Fundació Docència i Recerca Mútua Terrassa (https://www.mutuaterrassa.com/es/fundacio-recerca-i-docencia), and Post-Residency Grant for Research FMT (3rd edition, 2018). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Mathers CD, Boerma T, Ma Fat D. Global and regional causes of death. Br Med Bull. (2009) 92:7–32. doi: 10.1093/bmb/ldp028
3. Garau J, Calbo E. Community-acquired pneumonia. Lancet. (2008) 371:455–8. doi: 10.1016/s0140-6736(08)60216-0
4. Burman LA, Norrby R, Trollfors B. Invasive pneumococcal infections: incidence, predisposing factors, and prognosis. Clin Infect Dis. (1985) 7:133–42. doi: 10.1093/clinids/7.2.133
5. Rosen JB, Thomas AR, Lexau CA, Reingold A, Hadler JL, Harrison LH, et al. Geographic variation in invasive pneumococcal disease following pneumococcal conjugate vaccine introduction in the United States. Clin Infect Dis. (2011) 53:137–43. doi: 10.1093/cid/cir326
6. Miller E, Andrews NJ, Waight PA, Slack MP, George RC. Herd immunity and serotype replacement 4 years after seven-valent pneumococcal conjugate vaccination in England and Wales: an observational cohort study. Lancet Infect Dis. (2011) 11:760–8. doi: 10.1016/S1473-3099(11)70090-1
7. Helferty M, Rotondo JL, Martin I, Desai S. The epidemiology of invasive pneumococcal disease in the Canadian North from 1999 to 2010. Int J Circumpolar Health. (2013) 72:21606. doi: 10.3402/ijch.v72i0.21606
8. Elberse KE, van der Heide HG, Witteveen S, van de Pol I, Schot CS, van der Ende A, et al. Changes in the composition of the pneumococcal population and in IPD incidence in The Netherlands after the implementation of the 7-valent pneumococcal conjugate vaccine. Vaccine. (2012) 30:7644–51. doi: 10.1016/j.vaccine.2012.04.021
9. Link-Gelles R, Thomas A, Lynfield R, Petit S, Schaffner W, Harrison L, et al. Geographic and temporal trends in antimicrobial nonsusceptibility in streptococcus pneumoniae in the post-vaccine era in the United States. J Infect Dis. (2013) 208:1266–73. doi: 10.1093/infdis/jit315
10. Kozakova J, Krizova P, Maly M. Impact of pneumococcal conjugate vaccine on invasive pneumococcal disease in children under 5 years of age in the Czech Republic. PLoS ONE. (2021) 16:e0247862. doi: 10.1371/journal.pone.0247862
11. Severiche-Bueno DF, Severiche-Bueno DF, Bastidas A, Caceres EL, Silva E, Lozada J, et al. Burden of invasive pneumococcal disease (IPD) over a 10-year period in Bogotá, Colombia. Int J Infect Dis. (2021) 105:32–9. doi: 10.1016/j.ijid.2021.02.031
12. Sangil A, Xercavins M, Rodriguez-Carballeira M, Andres M, Riera M, Espejo E, et al. Impact of vaccination on invasive pneumococcal disease in adults with focus on the immunosuppressed. J Infect. (2015) 71:422–7. doi: 10.1016/j.jinf.2015.07.004
13. Naucler P, Darenberg J, Morfeldt E, Ortqvist A, Henriques Normark B. Contribution of host, bacterial factors and antibiotic treatment to mortality in adult patients with bacteraemic pneumococcal pneumonia. Thorax. (2013) 68:571–9. doi: 10.1136/thoraxjnl-2012-203106
14. Sangil A, Arranz MJ, Guerri-Fernandez R, Perez M, Monzon H, Payeras A, et al. Genetic susceptibility to invasive pneumococcal disease. Infect Genet Evol. (2018) 59:126–31. doi: 10.1016/j.meegid.2018.01.024
15. Calbo E, Garau J. Of mice and men: innate immunity in pneumococcal pneumonia. Int J Antimicrob Agents. (2010) 35:107–13. doi: 10.1016/j.ijantimicag.2009.10.002
16. Weinberger DM, Harboe ZB, Sanders EA, Ndiritu M, Klugman KP, Ruckinger S, et al. Association of serotype with risk of death due to pneumococcal pneumonia: a meta-analysis. Clin Infect Dis. (2010) 51:692–9. doi: 10.1086/655828
17. Aspa J, Rajas O, Rodriguez de Castro F, Huertas MC, Borderias L, Cabello FJ, et al. Impact of initial antibiotic choice on mortality from pneumococcal pneumonia. Eur Respir J. (2006) 27:1010–9. doi: 10.1183/09031936.06.00126004
18. Janoff EN, Musher DM. Streptococcus pneumoniae. In: Elsevier I, editor. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 2. 9th ed. (2020). p. 2473–91.
19. Alanee SR, McGee L, Jackson D, Chiou CC, Feldman C, Morris AJ, et al. Association of serotypes of Streptococcus pneumoniae with disease severity and outcome in adults: an international study. Clin Infect Dis. (2007) 45:46–51. doi: 10.1086/518538
20. Kalin M, Ortqvist A, Almela M, Aufwerber E, Dwyer R, Henriques B, et al. Prospective study of prognostic factors in community-acquired bacteremic pneumococcal disease in 5 countries. J Infect Dis. (2000) 182:840–7. doi: 10.1086/315760
21. French N, Gordon SB, Mwalukomo T, White SA, Mwafulirwa G, Longwe H, et al. A trial of a 7-valent pneumococcal conjugate vaccine in HIV-infected adults. N Engl J Med. (2010) 362:812–22. doi: 10.1056/NEJMoa0903029
22. Ludwig G, Garcia-Garcia S, Lanaspa M, Ciruela P, Esteva C, Fernandez de Sevilla M, et al. Serotype and clonal distribution dynamics of invasive pneumococcal strains after PCV13 introduction (2011–2016): surveillance data from 23 sites in Catalonia, Spain. PLoS ONE. (2020) 15:e0228612. doi: 10.1371/journal.pone.0228612
23. Masomian M, Ahmad Z, Ti Gew L, Poh CL. Development of Next Generation Streptococcus pneumoniae Vaccines Conferring Broad Protection. Vaccines. (2020) 8:132. doi: 10.3390/vaccines8010132
24. Lepoutre A, Varon E, Georges S, Dorleans F, Janoir C, Gutmann L, et al. Impact of the pneumococcal conjugate vaccines on invasive pneumococcal disease in France, 2001–2012. Vaccine. (2015) 33:359–66. doi: 10.1016/j.vaccine.2014.11.011
25. Munoz-Almagro C, Jordan I, Gene A, Latorre C, Garcia-Garcia JJ, Pallares R. Emergence of invasive pneumococcal disease caused by nonvaccine serotypes in the era of 7-valent conjugate vaccine. Clin Infect Dis. (2008) 46:174–82. doi: 10.1086/524660
26. Feikin DR, Kagucia EW, Loo JD, Link-Gelles R, Puhan MA, Cherian T, et al. Serotype-specific changes in invasive pneumococcal disease after pneumococcal conjugate vaccine introduction: a pooled analysis of multiple surveillance sites. PLoS Med. (2013) 10:e1001517. doi: 10.1371/journal.pmed.1001517
27. Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, et al. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. (2010) 201:32–41. doi: 10.1086/648593
28. Harboe ZB, Dalby T, Weinberger DM, Benfield T, Molbak K, Slotved HC, et al. Impact of 13-valent pneumococcal conjugate vaccination in invasive pneumococcal disease incidence and mortality. Clin Infect Dis. (2014) 59:1066–73. doi: 10.1093/cid/ciu524
29. von Gottberg A, de Gouveia L, Tempia S, Quan V, Meiring S, von Mollendorf C, et al. Effects of vaccination on invasive pneumococcal disease in South Africa. N Engl J Med. (2014) 371:1889–99. doi: 10.1056/NEJMoa1401914
30. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
31. Brouwer MC, de Gans J, Heckenberg SGB, Zwinderman AH, van der Poll T, van de Beek D. Host genetic susceptibility to pneumococcal and meningococcal disease: a systematic review and meta-analysis. Lancet Infect Dis. (2009) 9:31–44. doi: 10.1016/s1473-3099(08)70261-5
32. Kloek AT, Brouwer MC, van de Beek D. Host genetic variability and pneumococcal disease: a systematic review and meta-analysis. BMC Med Genomics. (2019) 12:130. doi: 10.1186/s12920-019-0572-x
33. Bogaert D, de Groot R, Hermans PWM. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. (2004) 4:144–54. doi: 10.1016/s1473-3099(04)00938-7
34. Hausdorff WP, Bryant J, Paradiso PR, Siber GR. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin Infect Dis. (2000) 30:100–21. doi: 10.1086/313608
35. Brueggemann AB, Peto TE, Crook DW, Butler JC, Kristinsson KG, Spratt BG. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J Infect Dis. (2004) 190:1203–11. doi: 10.1086/423820
36. Sjöström K, Spindler C, Ortqvist A, Kalin M, Sandgren A, Kuhlmann-Berenzon S, et al. Clonal and capsular types decide whether pneumococci will act as a primary or opportunistic pathogen. Clin Infect Dis. (2006) 42:451–9. doi: 10.1086/499242
37. Jansen AG, Rodenburg GD, van der Ende A, van Alphen L, Veenhoven RH, Spanjaard L, et al. Invasive pneumococcal disease among adults: associations among serotypes, disease characteristics, and outcome. Clin Infect Dis. (2009) 49:e23–9. doi: 10.1086/600045
38. Harboe ZB, Thomsen RW, Riis A, Valentiner-Branth P, Christensen JJ, Lambertsen L, et al. Pneumococcal serotypes and mortality following invasive pneumococcal disease: a population-based cohort study. PLoS Med. (2009) 6:e1000081. doi: 10.1371/journal.pmed.1000081
39. Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, Spratt BG. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis. (2003) 187:1424–32. doi: 10.1086/374624
40. Yu VL, Chiou CC, Feldman C, Ortqvist A, Rello J, Morris AJ, et al. An international prospective study of pneumococcal bacteremia: correlation with in vitro resistance, antibiotics administered, and clinical outcome. Clin Infect Dis. (2003) 37:230–7. doi: 10.1086/377534
41. Lujan M, Gallego M, Fontanals D, Mariscal D, Rello J. Prospective observational study of bacteremic pneumococcal pneumonia: Effect of discordant therapy on mortality. Crit Care Med. (2004) 32:625–31. doi: 10.1097/01.ccm.0000114817.58194.bf
42. Weinstein MP, Klugman KP, Jones RN. Rationale for revised penicillin susceptibility breakpoints versus Streptococcus pneumoniae: coping with antimicrobial susceptibility in an era of resistance. Clin Infect Dis. (2009) 48:1596–600. doi: 10.1086/598975
43. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. (2016) 164:337–40. doi: 10.1016/j.cell.2016.01.013
44. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. (2012) 336:1268–73. doi: 10.1126/science.1223490
45. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. (2014) 20:642–7. doi: 10.1038/nm.3568
46. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336:489–93. doi: 10.1126/science.1219328
47. Zhu B, Wang X, Li L. Human gut microbiome: the second genome of human body. Protein Cell. (2010) 1:718–25. doi: 10.1007/s13238-010-0093-z
48. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. (2017) 15:259–70. doi: 10.1038/nrmicro.2017.14
49. Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. (2012) 78:626–70. doi: 10.1128/aem.01051-12
50. Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. (2016) 535:85–93. doi: 10.1038/nature18849
51. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. (2013) 14:685–90. doi: 10.1038/ni.2608
52. Jochum L, Stecher B. Label or concept – what is a pathobiont? Trends Microbiol. (2020) 28:789–92. doi: 10.1016/j.tim.2020.04.011
53. Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2004) 170:266–72. doi: 10.1164/rccm.200403-354OC
54. Murphy TF, Brauer AL, Eschberger K, Lobbins P, Grove L, Cai X, et al. Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2008) 177:853–60. doi: 10.1164/rccm.200709-1413OC
55. Segal LN, Blaser MJ. A brave new world: the lung microbiota in an era of change. Ann Am Thoracic Soc. (2014) 11(Suppl 1):S21–S7. doi: 10.1513/annalsats.201306-189mg
56. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. (2011) 184:957–63. doi: 10.1164/rccm.201104-0655oc
57. Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. (2013) 1:19. doi: 10.1186/2049-2618-1-19
58. Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest. (1997) 111–1266–72. doi: 10.1378/chest.111.5.1266
59. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial topography of the healthy human lower respiratory tract. mBio. (2017) 8:e02287–16. doi: 10.1128/mbio.02287-16
60. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. (2011) 184(8):957–63. Epub 2011/06/18. doi: 10.1164/rccm.201104-0655OC
61. Man WH, van Houten MA, Mérelle ME, Vlieger AM, Chu MLJN, Jansen NJG, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med. (2019) 7:417–26. doi: 10.1016/s2213-2600(18)30449-1
62. Segal LN, Rom WN, Weiden MD. Lung microbiome for clinicians. New discoveries about bugs in healthy and diseased lungs. Ann Am Thorac Soc. (2014) 11:108–16. doi: 10.1513/AnnalsATS.201310-339FR
63. Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. (2014) 190:1283–92. doi: 10.1164/rccm.201407-1240OC
64. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
65. Relman DA. The human microbiome: ecosystem resilience and health. Nutr Rev. (2012) 70(Suppl 1):S2–9. doi: 10.1111/j.1753-4887.2012.00489.x
66. Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann Am Thorac Soc. (2015) 12(Suppl 2):S150–6. doi: 10.1513/AnnalsATS.201503-133AW
67. Bingula R, Filaire M, Radosevic-Robin N, Bey M, Berthon JY, Bernalier-Donadille A, et al. Desired turbulence? gut-lung axis, immunity, and lung cancer. J Oncol. (2017) 2017:5035371. doi: 10.1155/2017/5035371
68. Schuijt TJ, Lankelma JM, Scicluna BP, De Sousa E, Melo F, Roelofs JJTH, De Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–83. doi: 10.1136/gutjnl-2015-309728
69. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. (2014) 159:789–99. doi: 10.1016/j.cell.2014.09.053
70. Camelo-Castillo A, Henares D, Brotons P, Galiana A, Rodriguez JC, Mira A, et al. Nasopharyngeal microbiota in children with invasive pneumococcal disease: identification of bacteria with potential disease-promoting and protective effects. Front Microbiol. (2019) 10:11. doi: 10.3389/fmicb.2019.00011
71. Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio. (2016) 7:e01725–15. doi: 10.1128/mbio.01725-15
72. Lee DC, Kananurak A, Tran MT, Connolly PA, Polage CR, Iwase T, et al. Bacterial colonization of the hospitalized newborn: competition between staphylococcus aureus and staphylococcus epidermidis. Pediatr Infect Dis J. (2019) 38:682–6. doi: 10.1097/inf.0000000000002285
73. Zhou Y, Lin P, Li Q, Han L, Zheng H, Wei Y, et al. Analysis of the microbiota of sputum samples from patients with lower respiratory tract infections. Acta Biochim Biophys Sin. (2010) 42:754–61. doi: 10.1093/abbs/gmq081
74. Bousbia S, Papazian L, Saux P, Forel JM, Auffray JP, Martin C, et al. Repertoire of intensive care unit pneumonia microbiota. PLoS ONE. (2012) 7:e32486. doi: 10.1371/journal.pone.0032486
75. Iwai S, Fei M, Huang D, Fong S, Subramanian A, Grieco K, et al. Oral and airway microbiota in HIV-infected pneumonia patients. J Clin Microbiol. (2012) 50:2995–3002. doi: 10.1128/JCM.00278-12
76. Chaban B, Albert A, Links MG, Gardy J, Tang P, Hill JE. Characterization of the upper respiratory tract microbiomes of patients with pandemic H1N1 influenza. PLoS ONE. (2013) 8:e69559. doi: 10.1371/journal.pone.0069559
77. Leung RK, Zhou JW, Guan W, Li SK, Yang ZF, Tsui SK. Modulation of potential respiratory pathogens by pH1N1 viral infection. Clin Microbiol Infect. (2013) 19:930–5. doi: 10.1111/1469-0691.12054
78. de Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. (2016) 10:97–108. doi: 10.1038/ismej.2015.99
79. Chapman TJ, Morris MC, Xu L, Pichichero ME. Nasopharyngeal colonization with pathobionts is associated with susceptibility to respiratory illnesses in young children. PLOS ONE. (2020) 15:e0243942. doi: 10.1371/journal.pone.0243942
80. Rueca M, Fontana A, Bartolini B, Piselli P, Mazzarelli A, Copetti M, et al. Investigation of Nasal/oropharyngeal microbial community of covid-19 patients by 16S rDNA sequencing. Int J Environ Res Public Health. (2021) 18:2174. doi: 10.3390/ijerph18042174
81. Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. (2011) 6:e17035. doi: 10.1371/journal.pone.0017035
82. Mortensen MS, Brejnrod AD, Roggenbuck M, Abu Al-Soud W, Balle C, Krogfelt KA, et al. The developing hypopharyngeal microbiota in early life. Microbiome. (2016) 4:70. doi: 10.1186/s40168-016-0215-9
83. Bosch A, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Biesbroek G, Kool J, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med. (2017) 196:1582–90. doi: 10.1164/rccm.201703-0554OC
84. Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. (2017) 23:314–26. doi: 10.1038/nm.4272
85. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
86. Cremers AJ, Zomer AL, Gritzfeld JF, Ferwerda G, Van Hijum SA, Ferreira DM, et al. The adult nasopharyngeal microbiome as a determinant of pneumococcal acquisition. Microbiome. (2014) 2:44. doi: 10.1186/2049-2618-2-44
87. Paudel KR, Dharwal V, Patel VK, Galvao I, Wadhwa R, Malyla V, et al. Role of lung microbiome in innate immune response associated with chronic lung diseases. Front Med. (2020) 7:554. doi: 10.3389/fmed.2020.00554
88. Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global Strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report: gold executive summary. Arch Bronconeumol. (2017) 53:128–49. doi: 10.1016/j.arbres.2017.02.001
89. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. (2010) 5:e8578. doi: 10.1371/journal.pone.0008578
90. Wang Z, Singh R, Miller BE, Tal-Singer R, Van Horn S, Tomsho L, et al. Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: an analysis of the COPDMAP study. Thorax. (2018) 73:331–8. doi: 10.1136/thoraxjnl-2017-210741
91. Leitao Filho FS, Alotaibi NM, Ngan D, Tam S, Yang J, Hollander Z, et al. Sputum microbiome is associated with 1-year mortality after chronic obstructive pulmonary disease hospitalizations. Am J Respir Crit Care Med. (2019) 199:1205–13. doi: 10.1164/rccm.201806-1135OC
92. Zhao H, Liu J, Zhu J, Yang F, Wu H, Ba Y, et al. Bacterial composition and community structure of the oropharynx of adults with asthma are associated with environmental factors. Microb Pathog. (2020) 149:104505. doi: 10.1016/j.micpath.2020.104505
93. Durack J, Huang YJ, Nariya S, Christian LS, Ansel KM, Beigelman A, et al. Bacterial biogeography of adult airways in atopic asthma. Microbiome. (2018) 6:104. doi: 10.1186/s40168-018-0487-3
94. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. (2020) 323:1061–9. doi: 10.1001/jama.2020.1585
95. Soltani S, Zakeri A, Zandi M, Kesheh MM, Tabibzadeh A, Dastranj M, et al. The role of bacterial and fungal human respiratory microbiota in covid-19 patients. BioMed Res Int. (2021) 2021:1–13. doi: 10.1155/2021/6670798
96. Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, et al. Genomic diversity of severe acute respiratory syndrome–coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. (2020) 71:713–20. doi: 10.1093/cid/ciaa203
97. Gao Z, Kang Y, Yu J, Ren L. Human pharyngeal microbiome may play a protective role in respiratory tract infections. Genomics Proteomics Bioinformatics. (2014) 12:144–50. doi: 10.1016/j.gpb.2014.06.001
98. Wang Y, Li X, Ge T, Xiao Y, Liao Y, Cui Y, et al. Probiotics for prevention and treatment of respiratory tract infections in children: a systematic review and meta-analysis of randomized controlled trials. Medicine. (2016) 95:e4509. doi: 10.1097/MD.0000000000004509
99. Wilson MR, Zha L, Balskus EP. Natural product discovery from the human microbiome. J Biol Chem. (2017) 292:8546–52. doi: 10.1074/jbc.r116.762906
100. Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. (2016) 535:511–6. doi: 10.1038/nature18634
101. Manti S, Parisi GF, Papale M, Licari A, Salpietro C, Miraglia Del Giudice M, et al. Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for treatment of upper respiratory tract infections in children: a pilot study on short-term efficacy. Italian J Pediatr. (2020) 46:42. doi: 10.1186/s13052-020-0798-4
102. Guo Z, Lin H, Ji X, Yan G, Lei L, Han W, et al. Therapeutic applications of lytic phages in human medicine. Microb Pathog. (2020) 142:104048. doi: 10.1016/j.micpath.2020.104048
103. Van Der Kamp I, Draper LA, Smith MK, Buttimer C, Ross RP, Hill C. A new phage lysin isolated from the oral microbiome targeting streptococcus pneumoniae. Pharmaceuticals. (2020) 13:478. doi: 10.3390/ph13120478
104. Vazquez R, Garcia P. Synergy between two chimeric lysins to kill streptococcus pneumoniae. Front Microbiol. (2019) 10:1251. doi: 10.3389/fmicb.2019.01251
Keywords: microbiota, nasopharynx, Streptococcus pneumoniae, pneumococcal infections, invasive pneumococcal disease
Citation: Dietl B, Henares D, Boix-Palop L, Muñoz-Almagro C, Garau J and Calbo E (2021) Related Factors to Streptococcus pneumoniae Invasive Infection and Clinical Manifestations: The Potential Role of Nasopharyngeal Microbiome. Front. Med. 8:650271. doi: 10.3389/fmed.2021.650271
Received: 06 January 2021; Accepted: 22 March 2021;
Published: 20 April 2021.
Edited by:
Alessandro Russo, University of Pisa, ItalyReviewed by:
Salvatore Leonardi, University of Catania, ItalyLinghua Li, Guangzhou Eighth People's Hospital, China
Copyright © 2021 Dietl, Henares, Boix-Palop, Muñoz-Almagro, Garau and Calbo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esther Calbo, ecalbo@mutuaterrassa.es
 Beatriz Dietl
Beatriz Dietl Desirée Henares
Desirée Henares Lucía Boix-Palop1,2
Lucía Boix-Palop1,2  Carmen Muñoz-Almagro
Carmen Muñoz-Almagro Javier Garau
Javier Garau Esther Calbo
Esther Calbo