- 1Guangdong Institute of Microbiology, State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangzhou, China
- 2School of Bioscience and Bioengineering, South China University of Technology, Guangzhou, China
Eighty Listeria monocytogenes isolates were obtained from Chinese retail ready-to-eat (RTE) food and were previously characterized with serotyping and antibiotic susceptibility tests. The aim of this study was to characterize the subtype and virulence potential of these L. monocytogenes isolates by multilocus sequence typing (MLST), virulence-associate genes, epidemic clones (ECs), and sequence analysis of the important virulence factor: internalin A (inlA). The result of MLST revealed that these L. monocytogenes isolates belonged to 14 different sequence types (STs). With the exception of four new STs (ST804, ST805, ST806, and ST807), all other STs observed in this study have been associated with human listeriosis and outbreaks to varying extents. Six virulence-associate genes (inlA, inlB, inlC, inlJ, hly, and llsX) were selected and their presence was investigated using PCR. All strains carried inlA, inlB, inlC, inlJ, and hly, whereas 38.8% (31/80) of strains harbored the listeriolysin S genes (llsX). A multiplex PCR assay was used to evaluate the presence of markers specific to epidemic clones of L. monocytogenes and identified 26.3% (21/80) of ECI in the 4b-4d-4e strains. Further study of inlA sequencing revealed that most strains contained the full-length InlA required for host cell invasion, whereas three mutations lead to premature stop codons (PMSC) within a novel PMSCs at position 326 (GAA → TAA). MLST and inlA sequence analysis results were concordant, and different virulence potentials within isolates were observed. These findings suggest that L. monocytogenes isolates from RTE food in China could be virulent and be capable of causing human illness. Furthermore, the STs and virulence profiles of L. monocytogenes isolates have significant implications for epidemiological and public health studies of this pathogen.
Introduction
Listeria monocytogenes is a gram-positive, facultative intracellular bacterium that is responsible for listeriosis. Listeriosis can cause meningitis, newborn septicemia, encephalomyelitis, or even death in humans, especially in the elderly, pregnant women, or newborn. Every year, ~1591 cases of listeriosis in humans are report, with a 19% case-fatality rate in the United States (Scallan et al., 2011). In the European Union, a total of 1763 confirmed human cases of listeriosis (notification rate of 0.44 cases per 100,000 population) were reported in 2013 (EFSA, 2015). As an important foodborne pathogen, it is widespread in nature and lives naturally in plants and soil environments. Its ability to survive and grow over a wide range of environmental conditions, including refrigeration temperatures, high salt concentration and low pH, makes it a potential hazard in foods (Ryser and Marth, 2007).
Differences in virulence between L. monocytogenes strains may also influence infection and clinical outcome. Some strains are highly pathogenic and sometimes deadly, whereas others are less virulent or even avirulent and produce little harm in the host (Olier et al., 2002). Several methods have been used to differentiate L. monocytogenes strains. Based on somatic (O) and flagellar (H) antigens represents a conventional approach to understanding L. monocytogenes isolates ecological distribution and epidemiology. However, there are 13 serotypes and only four serotype (1/2a, 1/2b, 1/2c, and 4b) cause almost all cases of listeriosis in human. To further discriminate these strains, numerous molecular methods have been developed for epidemiological investigations and of help for the surveillance and control of listeriosis. Multilocus sequence typing (MLST) method is one of the most robust tools for investigating the global epidemiology of microbial populations (Sullivan et al., 2005). Based on the sequencing of seven housekeeping genes, it is highly discriminatory and provides unambiguous results that can be comparable directly among laboratories via the internet. In recent years, MLST has evolved to the reference method for global epidemiology and population biology (Maiden, 2006; Ragon et al., 2008).
Certain virulence and virulence-associated genes also play very important roles in intracellular survival, cell-to-cell spread, and virulence of L. monocytogenes. The presence of a number of virulence factors such as surface-associated internalins, listeriolysin O, and listeriolysin S (LLS) in L. monocytogenes significantly regulates its pathogenicity (Cotter et al., 2008; Shen et al., 2013). Of which, internalin A (InlA), encode by inlA, is responsible for facilitating the entry of L. monocytogenes into nonprofessional phagocytic cells expressing the human isoform of E-cadherin (Lecuit et al., 2001). It has been shown that mutations in inlA leading to a premature stop codon (PMSC) significantly reduce the invasion of the strain to human epithelial cells (Nightingale et al., 2005a). In addition, a small number of closely related L. monocytogenes clones have caused multiple outbreaks worldwide. These epidemic clones (EC) are divided into seven groups: ECI, ECII and ECIV within serotype 4b, ECIII, ECV and ECVII in serotype 1/2a, and ECVI within serotype 1/2b (Kathariou, 2003; Knabel et al., 2012; Lomonaco et al., 2013).
Since the main food linked to listeriosis outbreaks was ready-to-eat (RTE) food, the consumer had limit opportunities to destroy the pathogen before the food consumed. Therefore, the risk associated with strains isolated from RTE food may be more severe. In our previous study, 80 RTE L. monocytogenes isolates were examined (Wu et al., 2015a). In order to determine the potential pathogenic profile and relative risk of these RTE L. monocytogenes isolates, this study provide a phylogenetic framework based on MLST analysis of L. monocytogenes isolates and evaluate their virulence-genes and EC-specific markers. In addition, we analyzed the full-length sequences of important virulence factor (inlA) to investigate correlations among the MLST types, amino acid sequence of inlA, and virulence potential.
Materials and Methods
Bacterial Isolates
A total of 80 L. monocytogenes isolates were collected from retail ready-to-eat food samples in 24 Chinese cities were analyzed, comprising 27 isolates from cold vegetable dishes in sauce, 7 isolates from cold noodles dishes in sauce, 17 isolates from roast chicken, 12 isolates from roast duck, 11 isolates from roast pork, and 5 isolates from pasteurized milk (Table 1). The isolates were obtained between September 2012 and January 2014 according to the GB 4789.30-2010 of food microbiological examination of L. monocytogenes (National Food Safety Standards of China) with slight modifications and the most probable number (MPN) method (Gombas et al., 2003). They were identified by Gram stain, catalase, and oxidase tests and Micro ID Listeria identification system (Microgen, Camberley, UK). Serovars and antibiotic susceptibility were confirmed in previous study (Wu et al., 2015a). Additionally, 35 isolates were selected for the sequencing analysis of inlA, based on MLST profile, molecular serogroup, food origin, and antimicrobial resistance. Each isolate was incubated at 37°C overnight on TSB-YE (Tryptic soy agar with yeast extract). Genomic DNA was extracted using a Genomic DNA Extraction kit (Dongsheng Biotech, Guangzhou, China) according to the manufacturer's instructions. The concentration of genomic DNA was determined at 260 nm using a NanoDrop-ND-1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, MA, USA).
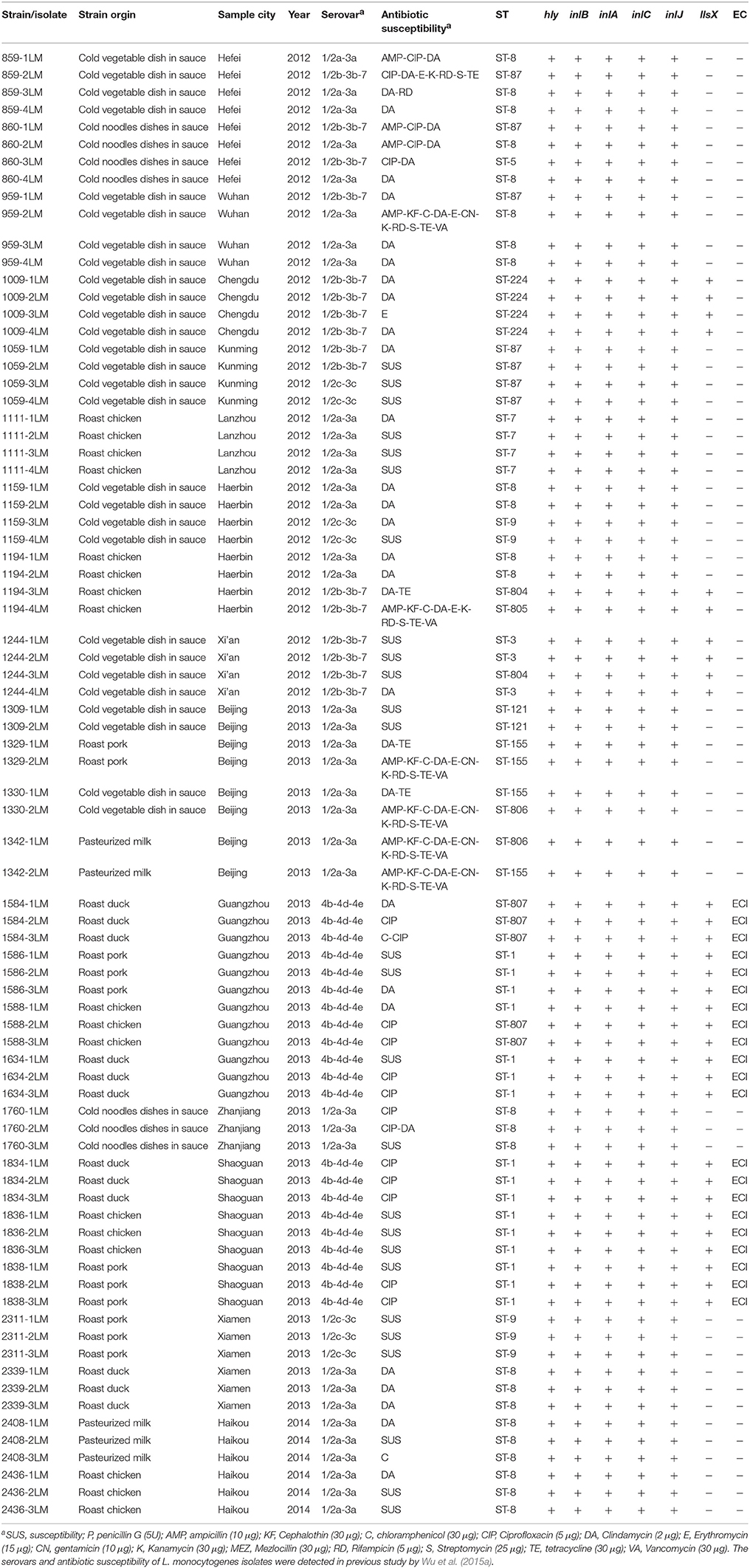
Table 1. Characteristics of 80 L. monocytogenes strains isolated from retail ready-to-eat food in this study.
Multilocus Sequence Typing
The MLST scheme used to characterize L. monocytogenes isolates is based on the sequence analysis of the following seven housekeeping genes: abcZ (ABC transporter), bglA (beta-glucosidase), cat (catalase), dapE (succinyl diaminopimelate desuccinylase), dat (D-amino acid aminotransferase), ldh (L-lactate dehydrogenase), and lhkA (histidine kinase) (Salcedo et al., 2003). The PCR amplification conditions were as follow: an initial cycle of 94°C for 4 min; 35 cycles of 94°C for 30 s, 52°C for 30 s (45°C for bglA), 72°C for 2 min, and a final extension at 72°C for 10 min. The DNA fragments were purified using a PCR purification kit (Qiagen, Genman) and were sequenced in each direction with Big Dye fluorescent terminators on an ABI3730XL sequencer (Applied BioSystems).
Determination of Virulence Genes and Epidemic Clone
The isolates were identified using duplex PCR detection containing hly (707 bp) and inlB (367 bp) genes, which are specific to L. monocytogenes, as previously described (Xu et al., 2009). Multiplex PCR (Liu et al., 2007) was used to determine the presence of the virulence genes inlA, inlC, and inlJ. The llsX gene was detected by PCR assays to identify the LLS-positive L. monocytogenes strains (Clayton et al., 2011). Presumptive major ECs (ECI, ECII, and ECIII) were found in the isolates as described previously (Chen and Knabel, 2007). All primers and PCR conditions are presented in Supplementary Table 1.
inlA Gene Sequencing
The 2400 bp long inlA gene was sequenced in 35 isolates representing the clonal diversity of L. monocytogenes. External primers were used to amplification covering the whole inlA ORF and internal primers for sequencing (Supplementary Table 2). The inlA sequences were assembled using Seqman (DNASTAR, Lasergene). Mutation types were determined according to the site of mutation that leads to PMSC in inlA (Nightingale et al., 2005a) and by comparing the obtained inlA sequence data to that of the L. monocytogenes EGDe reference strain (Glaser et al., 2001).
Data Analysis
For each MLST locus, an allele number was given to each distinct sequence variant, and a distinct sequence type (ST) number was attributed to each distinct combination of alleles among the seven genes. Sequence types (STs) were determined by using the L. monocytogenes MLST database (http://bigsdb.web.pasteur.fr/listeria/listeria.html). Sequence Type Analysis and Recombinational Tests software (S.T.A.R.T. ver.2; http://pubmlst.org/software/analysis/start2) was used to analyze the data of MLST.
Nucleotide diversity (π, average pairwise nucleotide difference/site; and k, average pairwise nucleotide difference/sequence), number of polymorphic sites, number of mutations, number of alleles, G+C content, Tajima's D test for neutrality, number of synonymous mutations (Ks), number of nonsynonymous mutations (Ka), and the Ks/Ka ratios [the number of nonsynonymous substitutions/nonsynonymous site (Ks) to the number of synonymous substitutions/synonymous site (Ka)] with a Jukes and Cantor correction were calculated using DnaSP version 5.10.01. Sequence analysis of inlA was performed using the ClustalX algorithm (version 1.83) (Thompson et al., 1997), which was followed by phylogenetic analysis using the maximum likelihood algorithm in MEGA 6 (version 6.05) (Tamura et al., 2011).
Results
Multilocus Sequence Typing of L. monocytogenes Isolates
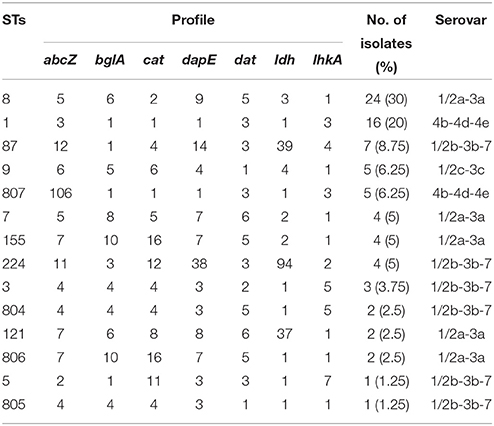
MLST detected a total of 14 different sequence types in the 80 isolates, including four new STs (ST804, ST805, ST806, and ST807) (Table 2). The most common allelic profile was ST8 (24/80, 30% of isolates) followed by ST1 (16/80, 20% of isolates) independently of the isolates source. With the exception of ST5 and ST805, the remainder of the STs included more than one isolate. Of the STs, ST-87, ST5, ST224, ST804, ST805, and ST3 belonged to serovar II.2 (1/2b-3b-7), ST8, ST7, ST122, ST155, and ST806 belonged to serovar I.1 (1/2a-3a), ST1 and ST807 belonged to serovars II.1 (4b-4d-4e), and ST9 belonged to serovar I.2 (1/2c-3c). Based on each ST matches at least one other ST at ≥6 of the 7 loci, three clonal complexes (CCs) were identified, including CC1 (ST1, ST807), CC3 (ST3, ST804), and CC155 (ST155, ST806). A phylogenetic tree based on the seven concatenated MLST sequences (Figure 1) shows the relatedness between the RTE strains. STs correlated well with serotype and lineage. Most isolates recovered from same city were clustered into one type. Furthermore, it should be noted that the multidrug resistant strains 1194-4LM, 1330-2LM, and 1342-1LM were belonged to new STs (ST805, ST806) indicating that these isolates were genetically diverse from other isolates.
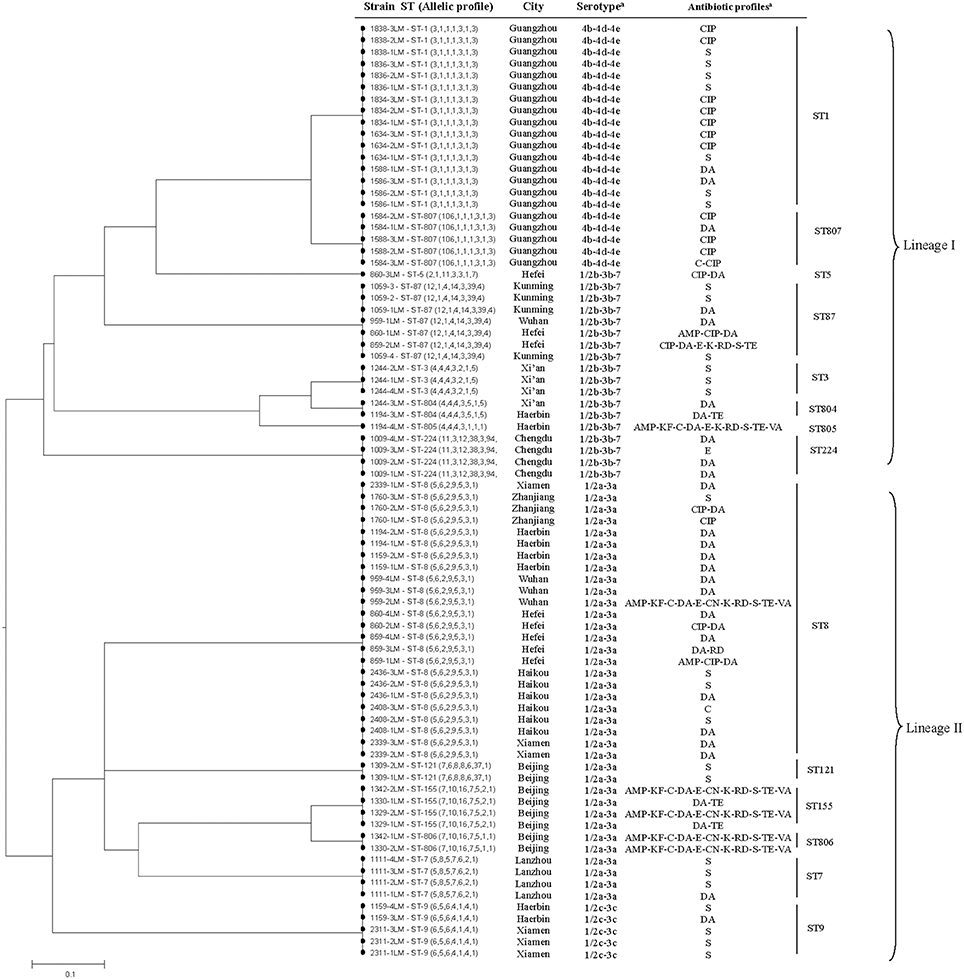
Figure 1. The UPGMA (unweighted pair group method with arithmetic mean) tree of the seven multi-locus sequence typing loci (3288 base pair concatenated length) of RTE L. monocytogenes isolates (Supplementary Table 1). This tree was generated using the S.T.A.R.T (version 2).
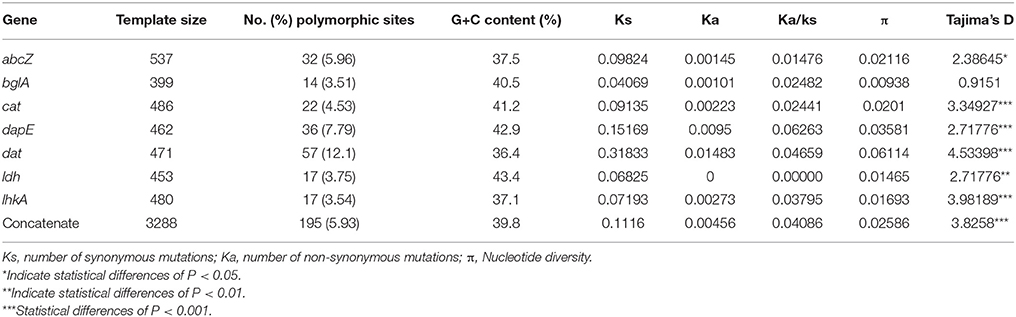
The evolutionary characteristics of the seven housekeeping genes among the isolates were analyzed using the software dnasp5, which can distinguish between randomly (i.e., neutrally) and non-randomly evolving DNA sequences. Tajima's D test indicated that abcZ, cat, dapE, dat, ldh, and lhkA evolved randomly, whereas bglA evolved non-significantly (0.9151, P > 0.10) (Table 3). However, seven gene portions harbored a total of 195 polymorphisms (5.93%; range 3.51–12.1% per gene). The average nucleotide diversity π was 2.6%, ranging from 0.9 to 6.2% per gene. The GC% observed in all alleles ranged from 36.4 to 42.9%, which was consistent with the 39% value observed across the entire L. monocytogenes EGDs genome (Glaser et al., 2001).
Prevalence of Virulence Associated Genes and EC Markers
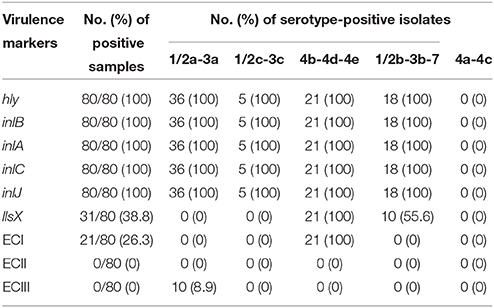
Eighty isolates of L. monocytogenes from retail RTE food were detected for the presence of virulence genes. The result showed that all isolates harbored listeriolysin O genes (hly) and internalin genes (inlA, inlB, inlC, and inlJ), and 38.8% (31/80) of isolates harbored listeriolysin S genes (llsX) (Table 4). The llsX-positive isolates including twenty-one 4b-4d-4e isolates, eleven 1/2b-3b-7 isolates. EC markers were identified in 26.3% (21/80) of isolates by multiplex PCR. ECI was the only EC marker identified and was observed in 4b-4d-4e isolates from roast meat (roast chicken/duck/pork) in this study. These EC isolates were also found to be positive for llsX.
Analysis of Selected Isolates for inlA Sequence
Sequencing the full 2400 bp inlA ORF in 35 L. monocytogenes isolates yielded 14 different inlA allelic types, indicating 88.6% of gene diversity. A total of 126 (5.2%) sites were polymorphic, including 85 sites with synonymous substitutions and 41 sites with non-synonymous substitutions. The average number of nucleotide differences per site between two sequences (π) was 0.01907; the average number of nucleotide differences (k) was 45.834. The GC% observed in inlA was 37.1%.
The phylogenetic tree generated using the inlA sequences of 35 RTE isolates and the L. monocytogenes EGDe reference strain (NC_003210.1; serotype 1/2a, ST35) revealed a similar grouping based on MLST analysis (Figure 2). The same lineage exhibited close relationships. Truncated forms of InlA have been described and are associated with reduced virulence (Nightingale et al., 2005a; Van Stelten et al., 2010), three distinct inlA alleles were found to harbor PMSC at position 492 (PMSC type 6), carried by ST 121 (1309-1LM, 1/2a-3a), the mutation at position 685 (PMSC type 11) harbored by the serotype 1/2c-3c isolate (2311-1LM, ST9) and a nonsense mutation at position 326 (GAA → TAA) of the inlA gene where a change of glutamic acid codon to a stop codon occurs showed a novel mutation type of PMSC (1159-3LM, 1/2c-3c, ST9).
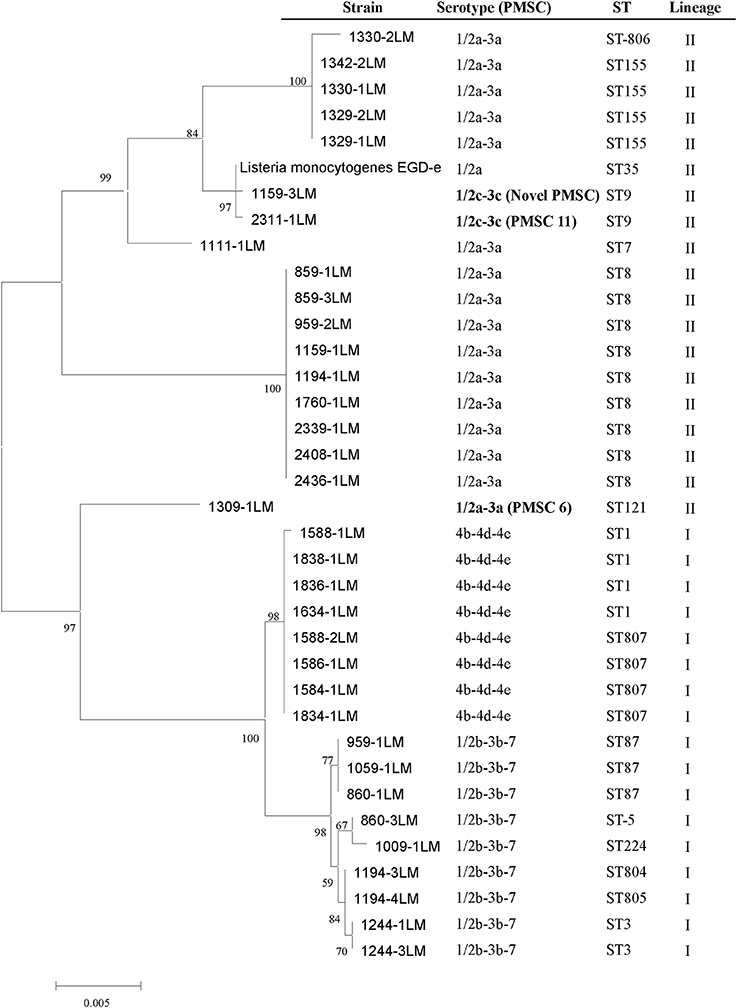
Figure 2. Maximum likelihood tree of selected RTE L. monocytogenes isolates based on inlA sequence. The inlA sequences of 36 isolates are shown, including 35 selected isolates in this study and Listeria monocytogenes EGDe.
Comparison of the amino-acid sequences of inlA between 35 L. monocytogenes isolates and the homologous sequence type strain EGDs, 13 internalin A variants were grouped. Each STs showed same variants, excluding CC1 (ST1, ST807), CC3 (ST3, ST804), and ST5 and ST805. Isolates from ST1 divided into two variants. In total, 40 amino acids were substituted (40/800, 5%). Most of the substitutions (31/40, 77.5%) occurred in the leucine-rich repeats (LLR), the inter-repeat (IR) domain and the B-repeat domain (Figure 3). The most conserved region of InlA among the 35 RTE L. monocytogenes isolates was the LRR domain. In parallel, a previous study (Ragon et al., 2008) reported the high constraint of the LRR-region and the moderate constraint of the IR- and B-repeat regions.
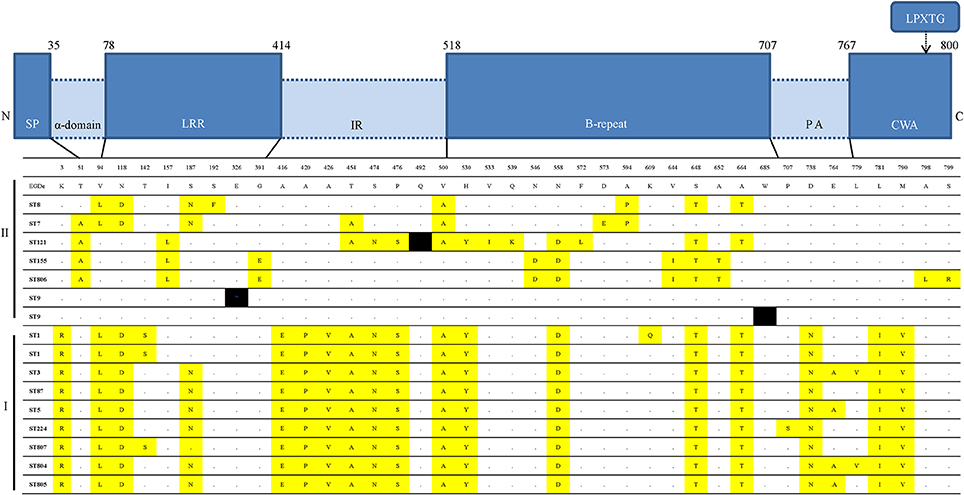
Figure 3. Internalin A variants in the 16 distinct ST and inlA alleles encountered. Comparison of the amino acid sequences of inlA of 35 L. monocytogenes isolated from Chinese RTE food to the homologous sequence of type strain EGDs. InlA functional domains are represented as distinct blocks: SP, signal peptide, α-domain, alpha-domain linker; LRRs, leucine rich repeats; IR, intragenic repeat, B-repeats, PA, pre-anchor domain; CWA, cell wall anchor; and C, C terminus. Yellow represents replacement with amino-acid and black indicates nonsense mutations leading to a premature stop codon (PMSC).
Discussion
As an important foodborne pathogen, L. monocytogenes remains a signification public health and food safety threat worldwide. In China, many studies of the prevalence of L. monocytogenes in food have been reported (Chen et al., 2009, 2014), but the outbreaks and infection of listeriosis in food are very limited. However, not all L. monocytogenes isolates have an equal capacity to cause disease. Therefore, it is important to investigate the molecular characteristics and virulence potential of L. monocytogenes isolates for designing and implementing more effective prevention strategies. The RTE isolates analyzed in this study were isolated from most of provincial capitals of China (Wu et al., 2015a), which could be better understanding the characterization of this important pathogen in China.
To enable increased characterization, MLST can be complemented by the nucleotide sequence determination of one or more highly diverse genes, such as those encoding antigens or antibiotic resistance determinants (Sullivan et al., 2005). In this study, 80 L. monocytogenes RTE isolates belonged to limited number of major clones. Except four new STs (ST804, 805, 806, 807), the other detected STs were already described in the L. monocytogenes Institute Pasteur MLST database (http://bigsdb.web.pasteur.fr/perl/bigsdb/bigsdb.pl?db=pubmlst_listeria_isolates_public&page=profiles): 327 strains of ST1 (51% from human isolates), 253 strains of ST3 (37% from human and 55% from food and environment), 143 strains of ST7 (63% from animal and feed), 37 strains of ST8 (including 15 human and 10 food), 130 strains of ST9 (35% from human and 21% from food), 52 strains of ST5 (50% from human and 23% from food), 22 strains of ST87 (55% from human and 23% from food), 70 strains of ST155 (50% from human), 6 strains of ST224 (2 isolates from human and 4 from environment), and 74 strains of ST121 (54% from food). Strains of all STs observed in our study have been associated to various extents with human listeriosis and outbreaks, indicating that L. monocytogenes strains of these STs have at least a theoretical pathogenic potential. It is worth nothing that ST1, comprising 20% of the isolates in this study, have the same sequences as F2365 (a serotype 4b isolates from a soft cheese outbreak in California in 1985), CLIP12848 (a serotype 4b isolates from outbreak in France in 1989), and CLIP68868 (a serotype 4b isolates from outbreak in Sweden 1995) which should the worthy of the attention of the food hygiene supervision department. Interestingly, novel STs (ST805, ST806) were found in multidrug resistant strains (Figure 1), showing some allele numbers had exchanged. Multiple resistance in L. monocytogenes was linked to the presence of a self-transferable plasmid that was proposed to originate in Enterococcus-Streptococcus (Charpentier and Courvalin, 1999). Therefore, the self-transferable plasmid may also impact the homologous recombination of housekeeping genes. However, further research is needed to determine the reason underlying this correlation.
The presence of hly, inlA, inlB, inlC, inlJ, and llsX was evaluated in L. monocytogenes isolates recovered from patients, food and the environment (Kathariou, 2003; Wieczorek et al., 2012; Wu et al., 2015b). In this study, PCR based analysis of the 80 RTE isolates showed that most of these isolates possessed virulence genes similar to those of clinical isolates (Mammina et al., 2009). The hly gene is encoding the listeriolysin O (LLO), the determinant that is required for the disruption of the phagocytic vacuole and the release of bacteria into the cytoplasm, a prerequisite for their intracellular proliferation. Therefore, LLO is an essential virulence factor and its absence leads to total avirulence (Portnoy et al., 1992). L. monocytogenes adheres to and actively through actions of internalins, a complex family of LRR-containing proteins (Bierne et al., 2007). In this study, we selected inlA, inlB, inlC, and inlJ for tested internalins in L. monocytogenes isolates. These genes are claimed to play a role in pathogenesis of human listeriosis. Besides, the llsX gene (encoding LLS) was employed as a genetic marker to detect L. monocytogenes with Listeria pathogenicity island 3 (LIPI-3) (Cotter et al., 2008), and strains from the present study exhibited 38.8% (31/80) positivity. The LLS, which is present in a subset of strains of lineage I, is induced only under oxidative stress conditions and contributes to murine virulence and in survival in polymorphonuclear neutrophils (Cotter et al., 2008). Overall, our result showed that the positive-llsX strains were found exclusively in lineage I (shown Table 3 and Figure 1), the evolutionary lineage of L. monocytogenes that contributes to the majority of spontaneous and epidemic outbreaks of listeriosis (Jeffers et al., 2001). Besides, most of positive-llsX strains involving ECI (21/32, 65.6%) have been related to several major outbreaks in the US and Europe in the past (Kathariou, 2003; Liu, 2008). In general, ECs is defined as a small number of isolates of a presumably common ancestor (Chen and Knabel, 2007). Specific ECs continue to be associated with sporadic cases around the world (Neves et al., 2008; Mammina et al., 2009; Lomonaco et al., 2013). Recently, Cantinelli et al. (2013) conclude that the “epidemic clone” denominations represent a redundant but largely incomplete nomenclature system for MLST-defined clones, which must be regarded as successful genetic groups that are widely distributed across time and space. In our study, L. monocytogenes isolates of MLST-CC1, including ST1 and ST807 were both ECI which was consistent with this conclusion. However, the high occurrence of multiple virulence-associated genes and ECI markers in L. monocytogenes isolates from Chinese RTE food could pose a significant health risk as these isolates can be pathogenic and potentially capable of causing an epidemic.
Certainly, the presence of these virulence-associated genes does not mean a particular strain is virulent, as these genes are normally present in L. monocytogenes but if absence of these genes lead to avirulence. Over the past decade, previous studies identified naturally occurring PMSC mutations in inlA and have demonstrated that these mutations are responsible for virulence attenuation (Van Stelten and Nightingale, 2008). L. monocytogenes strains with PMSCs were frequently isolated from RTE foods in the US, although inlA PMSCs were markedly underrepresented among ECs (Van Stelten et al., 2010). In our study, the sequencing of inlA genes from isolates revealed that three mutations resulted in PMSCs: type 6, 11 and a novel type at position 326 (GAA → TAA), which means most of strains (32/35, 91.4%) are capable of producing full-length InlA required for host cell invasion. Generally, PMSCs have been reported 30%-45% of prevalence in food but 5% of prevalence in clinical isolates (Jacquet et al., 2004; Van Stelten et al., 2010; Chen et al., 2011). Three mutations were both harbored by the lineage II strains (2 strains with ST9, one strain with ST121) (Figure 2), which further confirmed that 1/2a and 1/2c (lineage II) more frequently possess PMSCs than 1/2b or 3b serotypes, with 4b strains rarely having PMSCs (Orsi et al., 2011). PMSC seems to be more frequently observed in ST9 and ST121 isolates as previous reports (Ragon et al., 2008; Ciolacu et al., 2014). ST9 and ST121 include isolates from several different countries and from several different environmental, clinical and food source (Ragon et al., 2008; Parisi et al., 2010; Holch et al., 2013). Some L. monocytogenes strains can persist for a longtime in food processing. The repeatedly lost by convergent evolution in the genetically homogeneous of these genotypes, may be attribute to the selective advantage by the loss of a functional InlA protein or a relaxed selective constraint on maintaining InlA function (Ragon et al., 2008). It is worth mentioning that the novel PMSCs type at position 326 (GAA → TAA), indicated that variation occurred in the LLR domain were located in repeats 10 (Figure 3). This domain promotes interaction with human surface receptor, E-cadherin, and has been reported as highly conserved, especially from repeats 7 to 15 (Ragon et al., 2008). To date, at least 18 distinct mutations in inlA leading to PMSCs have been observed in L. monocytogenes isolates worldwide, only four of them (type 5, 16, 17, and 18) were found in LLR (Van Stelten et al., 2010). Compared with the inlA variants, L. monocytogenes retained the ability for localized recombination is clearly provided by the inlA gene coding InlA, in agreement with previous reports (Nightingale et al., 2005b; Orsi et al., 2007). The substitution of some amino acid were consistent between lineage I strains and II, such as lysine replaced into arginine at position 3 in all lineage I strains (Figure 3). Additionally, the close relationship between inlA sequencing and MLST typing (Figure 2) suggested that housekeeping genes and virulence-associated genes had some similar genetic characterization, representing monophyletic origin. In other words, our data suggest that L. monocytogenes evolved and generated different STs related to some degree of evolutionary consistent.
In summary, the research findings suggest that L. monocytogenes isolated from Chinese RTE food showed genetic relatedness and virulence attribute. Serotype, lineage, MLST types, even antibiotic resistance showed some degree of genetically relationship. Most MLST sequence types (10/14, 71.4%) found in this study have been linked to human listeriosis around the world. All isolates carried several important virulence genes (inlA, inlB, inlC, inlJ, and hly), whereas 38.8% (31/80) of strains harbored listeriolysin S genes (llsX). Besides, all 4b-4d-4e isolates belonged to ECI showed a high potential to cause human diseases. Further study of inlA sequencing showed most of selected strains (32/35, 91.4%) contained PMSC-lacking inlA gene sequences required for the encoding of InlA factor and bacterial invasion of the host cell. Based on the genetic properties observed in L. monocytogenes isolates, it is reasonable to assume L. monocytogenes found in Chinese retail RTE food have potential to cause listeriosis, and is concerning. These data have significant implications for the epidemiological and public health studies of this pathogen.
Author Contributions
Conceived and designed the experiments: QW, JZ, SW. Performed the experiments: SW. Analyzed the data: SW, MC. Contributed reagents/materials/analysis tools: WG. Contributed to the writing of the manuscript: SW, QW.
Funding
We would like to acknowledge the financing support of National Natural Science Foundation of China (No. 31471664), the Science and Technology Projects of Guangdong (2013B050800026), and the Science and Technology Projects of Guangzhou (201504010036).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RT and handling Editor declared a current collaboration and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00168
References
Bierne, H., Sabet, C., Personnic, N., and Cossart, P. (2007). Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9, 1156–1166. doi: 10.1016/j.micinf.2007.05.003
Cantinelli, T., Chenal-Francisque, V., Diancourt, L., Frezal, L., Leclercq, A., Wirth, T., et al. (2013). “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 51, 3770–3779. doi: 10.1128/JCM.01874-13
Charpentier, E., and Courvalin, P. (1999). Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 43, 2103–2108.
Chen, J., Zhang, X., Mei, L., Jiang, L., and Fang, W. (2009). Prevalence of Listeria in Chinese food products from 13 provinces between 2000 and 2007 and virulence characterization of Listeria monocytogenes isolates. Foodborne Pathog. Dis. 6, 7–14. doi: 10.1089/fpd.2008.0139
Chen, M., Wu, Q., Zhang, J., Yan, Z., and Wang, J. (2014). Prevalence and characterization of Listeria monocytogenes isolated from retail-level ready-to-eat foods in South China. Food Control 38, 1–7. doi: 10.1016/j.foodcont.2013.09.061
Chen, Y., and Knabel, S. J. (2007). Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 73, 6299–6304. doi: 10.1128/AEM.00961-07
Chen, Y., Ross, W. H., Whiting, R. C., Van Stelten, A., Nightingale, K. K., Wiedmann, M., et al. (2011). Variation in Listeria monocytogenes dose responses in relation to subtypes encoding a full-length or truncated internalin A. Appl. Environ. Microbiol. 77, 1171–1180. doi: 10.1128/AEM.01564-10
Ciolacu, L., Nicolau, A. I., Wagner, M., and Rychli, K. (2014). Listeria monocytogenes isolated from food samples from a Romanian black market show distinct virulence profiles. Int. J. Food Microbiol. 209, 44–51. doi: 10.1016/j.ijfoodmicro.2014.08.035
Clayton, E. M., Hill, C., Cotter, P. D., and Ross, R. P. (2011). Real-time PCR assay to differentiate listeriolysin S-positive and-negative strains of Listeria monocytogenes. Appl. Environ. Microbiol. 77, 163–171. doi: 10.1128/AEM.01673-10
Cotter, P. D., Draper, L. A., Lawton, E. M., Daly, K. M., Groeger, D. S., Casey, P. G., et al. (2008). Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4:e1000144. doi: 10.1371/journal.ppat.1000144
EFSA (2015). The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 13:3991. doi: 10.2903/j.efsa.2015.3991
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., et al. (2001). Comparative genomics of Listeria species. Science 294, 849–852. doi: 10.1126/science.1063447
Gombas, D. E., Chen, Y., Clavero, R. S., and Scott, V. N. (2003). Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66, 559–569.
Holch, A., Webb, K., Lukjancenko, O., Ussery, D., Rosenthal, B. M., and Gram, L. (2013). Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl. Environ. Microbiol. 79, 2944–2951. doi: 10.1128/AEM.03715-12
Jacquet, C., Doumith, M., Gordon, J. I., Martin, P. M., Cossart, P., and Lecuit, M. (2004). A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189, 2094–2100. doi: 10.1086/420853
Jeffers, G. T., Bruce, J. L., McDonough, P. L., Scarlett, J., Boor, K. J., and Wiedmann, M. (2001). Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147, 1095–1104. doi: 10.1099/00221287-147-5-1095
Kathariou, S. (2003). “Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes,” in Microbial Food Safety in Animal Agriculture: Current Topics, eds M. E. Torrence and R. E. Isaacson (Oxford, UK: Blackwell Publishing), 243–256. doi: 10.1002/9780470752616.ch25
Knabel, S. J., Reimer, A., Verghese, B., Lok, M., Ziegler, J., Farber, J., et al. (2012). Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 50, 1748–1751. doi: 10.1128/JCM.06185-11
Lecuit, M., Vandormael-Pournin, S., Lefort, J., Huerre, M., Gounon, P., Dupuy, C., et al. (2001). A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292, 1722–1725. doi: 10.1126/science.1059852
Liu, D., Lawrence, M. L., Austin, F. W., and Ainsworth, A. J. (2007). A multiplex PCR for species-and virulence-specific determination of Listeria monocytogenes. J. Microbiol. Methods 71, 133–140. doi: 10.1016/j.mimet.2007.08.007
Lomonaco, S., Verghese, B., Gerner-Smidt, P., Tarr, C., Gladney, L., Joseph, L., et al. (2013). Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg. Infect. Dis. 19, 147. doi: 10.3201/eid1901.121167
Maiden, M. C. (2006). Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60, 561–588. doi: 10.1146/annurev.micro.59.030804.121325
Mammina, C., Aleo, A., Romani, C., Pellissier, N., Nicoletti, P., Pecile, P., et al. (2009). Characterization of Listeria monocytogenes isolates from human listeriosis cases in Italy. J. Clin. Microbiol. 47, 2925–2930. doi: 10.1146/annurev.micro.59.030804.121325
Neves, E., Lourenço, A., Silva, A. C., Coutinho, R., and Brito, L. (2008). Pulsed-field gel electrophoresis (PFGE) analysis of Listeria monocytogenes isolates from different sources and geographical origins and representative of the twelve serovars. Syst. Appl. Microbiol. 31, 387–392. doi: 10.1016/j.syapm.2008.08.005
Nightingale, K. K., Windham, K., and Wiedmann, M. (2005b). Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187, 5537–5551. doi: 10.1128/JB.187.16.5537-5551.2005
Nightingale, K. K., Windham, K., Martin, K. E., Yeung, M., and Wiedmann, M. (2005a). Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71, 8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005
Olier, M. W., Pierre, F., Lemaître, J.-P., Divies, C., Rousset, A., and Guzzo, J. (2002). Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148, 1855–1862. doi: 10.1099/00221287-148-6-1855
Orsi, R. H., Ripoll, D. R., Yeung, M., Nightingale, K. K., and Wiedmann, M. (2007). Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiology 153, 2666–2678. doi: 10.1099/mic.0.2007/007310-0
Orsi, R. H., den Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Parisi, A., Latorre, L., Normanno, G., Miccolupo, A., Fraccalvieri, R., Lorusso, V., et al. (2010). Amplified fragment length polymorphism and multi-locus sequence typing for high-resolution genotyping of Listeria monocytogenes from foods and the environment. Food Microbiol. 27, 101–108. doi: 10.1016/j.fm.2009.09.001
Portnoy, D. A., Chakraborty, T., Goebel, W., and Cossart, P. (1992). Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60, 1263.
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Ryser, E. T., and Marth, E. H. (2007). Listeria, Listeriosis, and Food Safety. New York, NY: CRC Press.
Salcedo, C., Arreazá, L., Alcala, B., de la Fuente, L., and Vázquez, J. (2003). Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41, 757–762. doi: 10.1128/JCM.41.2.757-762.2003
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.p11101
Shen, J., Rump, L., Zhang, Y., Chen, Y., Wang, X., and Meng, J. (2013). Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiol. 35, 58–64. doi: 10.1016/j.fm.2013.02.014
Sullivan, C. B., Diggle, M. A., and Clarke, S. C. (2005). Multilocus sequence typing. Mol. Biotechnol. 29, 245–254. doi: 10.1385/MB:29:3:245
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Van Stelten, A., and Nightingale, K. (2008). Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 74, 7365–7375. doi: 10.1128/AEM.01138-08
Van Stelten, A., Simpson, J., Ward, T., and Nightingale, K. (2010). Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 76, 2783–2790. doi: 10.1128/AEM.02651-09
Wieczorek, K., Dmowska, K., and Osek, J. (2012). Prevalence, characterization, and antimicrobial resistance of Listeria monocytogenes isolates from bovine hides and carcasses. Appl. Environ. Microbiol. 78, 2043–2045. doi: 10.1128/AEM.07156-11
Wu, S., Wu, Q., Zhang, J., Chen, M., and Hu, H. (2015b). Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PLoS ONE 10:e0136682. doi: 10.1371/journal.pone.0136682
Wu, S., Wu, Q., Zhang, J., Chen, M., and Yan, Z. (2015a). Prevalence, antibiotic resistance and genetic diversity of Listeria monocytogenes isolated from retail ready-to-eat foods in China. Food Control 47, 340–347. doi: 10.1016/j.foodcont.2014.07.028
Keywords: Listeria monocytogenes, MLST, inlA, PMSC, virulence genes, epidemic clones
Citation: Wu S, Wu Q, Zhang J, Chen M and Guo W (2016) Analysis of Multilocus Sequence Typing and Virulence Characterization of Listeria monocytogenes Isolates from Chinese Retail Ready-to-Eat Food. Front. Microbiol. 7:168. doi: 10.3389/fmicb.2016.00168
Received: 20 October 2015; Accepted: 01 February 2016;
Published: 16 February 2016.
Edited by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyReviewed by:
Luca Settanni, Università degli Studi di Palermo, ItalyFatih Ozogul, Çukurova Üniversity, Turkey
Rosanna Tofalo, Università degli Studi di Teramo, Italy
Copyright © 2016 Wu, Wu, Zhang, Chen and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, wuqp203@163.com
 Shi Wu
Shi Wu Qingping Wu
Qingping Wu Jumei Zhang1
Jumei Zhang1 Moutong Chen
Moutong Chen