- 1Department of Veterinary Medicine and Public Health, College of Veterinary Medicine and Biomedical Sciences, Sokoine University of Agriculture, Morogoro, Tanzania
- 2Department of Fisheries Development, National Fish Quality Control Laboratory-Nyegezi, Mwanza, Tanzania
- 3Department of Veterinary and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
- 4Mzumbe University, Mzumbe, Tanzania
- 5School of Chemical and Biomedical Engineering, Nanyang Technological University, Singapore, Singapore
Extended-spectrum β-lactamase (ESBL)-producing bacteria constitute an emerging global health issue with food products being vehicles of transmission and the aquatic environments serving as potential reservoirs. This study aimed to characterize ESBL-producing Escherichia coli in Nile perch and water from Lake Victoria in Tanzania. A total of 180 samples of Nile perch and 60 water samples were screened for ESBL-producing E. coli on MacConkey agar supplemented with 2 μg/ml of cefotaxime and confirmed by blaCTX–M and blaTEM PCR. Antimicrobial resistance was determined by the disk diffusion method, and the ESBL-producing isolates were whole genome sequencing (WGS). ESBL-producing E. coli were detected in eight of the 180 analyzed Nile perch samples, and only one water sample was positive (1.7%, n = 60). Isolates were resistant to sulfamethoxazole–trimethoprim (100%), ampicillin/cloxacillin (100%), erythromycin 72.7% (8/11), tetracycline 90.9% (10/11), and nalidixic acid 63.6% (7/11). This mostly corroborates the resistance genes that they carried for sulfonamides (sul1 and sul2), trimethoprim (dfrA and dfrB), aminoglycosides [aac(3)-IId, strA, and strB], tetracycline [tet(B) and tet(D)], and fluoroquinolones (qepA4). They harbored plasmid replicon types IncF, IncX, IncQ, and Col and carried blaCTX–M–15 and blaTEM–1B genes generally found on the same contigs as the IncF plasmid replicon. Although epidemiologically unrelated, the strains formed three separate sequence type–phylogroup–serotype-specific clusters: C1, C2, and C3. Cluster C1 included five strains (3 to 13 SNPs) belonging to ST167, phylogroup A, and serotype O9:H21; the two C2 strains (11 SNPs) belong to ST156, phylogroup B1, and serotype ONT:H28; and C3 was made up of four strains (SNPs ranged from 4 to 17) of ST636, phylogroup B2, and serotype O45:H7. The common virulence gene gad was reported in all strains. In addition, strains in C2 and C3 possessed iss, lpfA, and nfaE virulence genes, and the vat gene was found only in C3. The present study reports the occurrence of multidrug-resistant ESBL-producing E. coli carrying plasmid-mediated ESBL genes in offshore water and Nile perch in Lake Victoria. Strains formed three clonal clusters of unknown origin. This study reveals that the Lake may serve as reservoir for ESBL-producing bacteria that can be transmitted by fish as a food chain hazard of One-Health concern.
Introduction
Escherichia coli and other related bacteria can produce extended-spectrum β-lactamase (ESBL) enzymes that hydrolyze a broad spectrum of β-lactam drugs such as cephalosporins (e.g., ceftazidime and cefotaxime) and monobactams (e.g., aztreonam and nocardicin), classes of antimicrobials that are critical in human medicine (Lavilla et al., 2008; Shaikh et al., 2015; Adelowo et al., 2018). ESBL-producing Gram-negative bacteria especially those producing cefotaximase-Munich (CTX-M) enzymes have emerged as important pathogens causing healthcare- and community-associated infections worldwide.
Studies in many different countries increasingly document how foods may be important sources of ESBL-producing E. coli, such as vegetables, poultry, pork, and other animal foods. In particular, poultry meat has been reported commonly to be associated with ESBL-producing E. coli globally (Overdevest, 2011; Chishimba et al., 2016; Nguyen et al., 2016; Falgenhauer et al., 2019; Projahn et al., 2019). Possible horizontal transfer of resistance genes from urban sewage and clinical isolates to bacteria associated with fish and aquatic recipients of wastewater has been documented (Kümmerer, 2009; Martinez, 2009; Jiang et al., 2013; Blaak et al., 2014).
The β-lactamase enzymes are derived from mutations in temoneira (TEM), sulfhydryl variable (SHV), and CTX-M genes located on bacterial plasmids or chromosomes (Ben Said et al., 2015; Legese et al., 2017; Adelowo et al., 2018). These genes can easily be horizontally transferred from one bacterial strain to another including across bacterial species (Lavilla et al., 2008). The SHV enzyme seems more dominant in Klebsiella pneumoniae among the Enterobacteriaceae (Rupp and Fey, 2003). The CTX-M enzymes are often the most common ESBLs produced by E. coli and have been isolated in human clinical, animal food, and environmental samples (Egea et al., 2012; Boonyasiri et al., 2014; Dib et al., 2018). On the other hand, the TEM enzymes especially of TEM-1 and TEM-2 are predominantly reported in E. coli and other members of Enterobacteriaceae, although TEM-1 is not considered as ESBLs. These TEM enzymes have been reported from Klebsiella spp. and Enterobacter spp. isolated in food, clinical, and environmental samples around the world as previously described (Smet et al., 2010; Delgado et al., 2016).
The reservoirs of ESBL-producing E. coli are warm-blooded animals including humans, which can transmit the bacteria to different environments, for example, aquatic environments, through fecal pollution (Jiang et al., 2012). Recently, studies have reported ESBL-producing E. coli in seafood such as shrimps, sardines, farmed fish, Nile tilapia (Oreochromis niloticus) as well as in frozen mackerel (Jiang et al., 2012; Moremi et al., 2016b; Nasreldin and Khaldoon, 2015; Dib et al., 2018). It has been documented that urban sewage, for example, from hospitals, is an important source of ESBL-producing enteric bacteria (Ojer-Usoz et al., 2013; Abgottspon et al., 2014). Thus, ESBL-producing bacteria in sewage and in runoff water from agricultural soil fertilized with livestock manure can enter the aquatic environment where antimicrobial resistance genes may be transferred horizontally to the ubiquitous bacterial flora, although the rate and health importance of such transfer are unknown.
In Tanzania, ESBL-producing E. coli have been isolated from different sources such as human specimens, foods, and aquatic environments (Moyo et al., 2010; Moremi et al., 2016b). Although ESBL-producing E. coli have been reported in humans in different regions of Tanzania (Moyo et al., 2010; Seni et al., 2016; Katakweba et al., 2018) and also in livestock and poultry (Katakweba et al., 2018), the importance of livestock and their meats as sources of ESBL-producing E. coli is unknown. ESBL-producing Enterobacteriaceae recovered from the aquatic environments and tilapia from Lake Victoria were previously characterized (Moremi et al., 2016b). This study was carried out with tilapia, which is a fish species commonly found in shallow water, which often is polluted by discharged human wastes of different sources mostly of fecal origin. Thus, a variety of ESBL-producing bacteria were found in the samples analyzed by Moremi et al. (2016b). Our study investigated the occurrence of ESBL-producing Enterobacteriaceae spp. along the chain from capture to market including water and Nile perch at offshore deep water fishing areas. We further applied whole genome sequencing (WGS), which provides important information about antimicrobial resistance and virulence genes in bacterial pathogens and is a powerful molecular tool in investigations of disease outbreaks.
The aim of this study was to determine the occurrence and genomic characteristics of ESBL-producing E. coli isolated in water at offshore fishing grounds and in Nile perch (Lates niloticus) from Lake Victoria, Tanzania.
Materials and Methods
Study Design, Sample Collection, and Processing
The study employed a cross-sectional design and was conducted from February to July 2017. A total of 240 samples of water and Nile perch were collected and analyzed for ESBL-producing Enterobacteriaceae, and the isolates were tested for antimicrobial resistance. This sample size was calculated based on the estimated prevalence of ESBL-producing Enterobacteriaceae spp. detected in other fish spp. from Lake Victoria reported by Moremi et al. (2016b) using the following formula:
Out of 240 samples, 180 were Nile perch and were collected from fishing grounds (n = 60), landing sites (n = 60), and domestic fish markets (n = 60). In addition, offshore water samples (n = 60) were collected and analyzed from fishing grounds. The size of fish sampled ranged between 1 and 2 kg by weight. The selection of Nile perch samples at markets was based on the availability of fish from vendors. At each market, 10 samples were collected from five retailers, with two fish purchased from each vendor. We did not register how many Nile perch were on sale at each market. Details on sample collection, transport, and processing are provided by Baniga et al. (2019). Nile perch samples were prepared by weighing intestines, gills, flesh, and fish surface mucus in a ratio of 1:9 to Buffered Peptone Water (BPW) (Oxoid Ltd., Hampshire, United Kingdom). Fish intestines, gills, and flesh (15- to 25-g volumes) were homogenized for 60 s in a stomacher (Seward 400, United Kingdom), whereas fish mucus was obtained and analyzed by massaging the fish surface in a sterile stomacher bag containing 225 ml of BPW. The weight of fish gills and intestine samples was not always the same because the weight varies based on the size of the fish; for example, a fish weighing 1 kg will have gills and intestines each weighing below 25 g and not less than 15 g. Stomacher bags used to obtain the surface mucus of fish were of 16063/0606 size, that is, Stomacher® Bags, Seward Genuine 16063/0606.1 The collection of fish mucus sample was done while fish was still intact before other subsamples, that is, flesh, gills, and intestines, were collected. All fish samples were processed following standard method (ISO 6887-3,2003). Twenty-five milliliters of each water sample was filtered using a standard filter membrane of 0.45 μl of pore size (Thermo Fisher Scientific, Waltham, MA, United States). After filtration, the filter membrane was torn into pieces using sterile forceps and then placed into 225 ml of BPW, which were shaken rigorously and then incubated at 37°C overnight for enrichment before plating out on the MacConkey agar. Thus, the limit of detection was 4 colony-forming units (cfu) ESBL-producing Escherichia coli per 100 ml. Water sample handling and processing were done according to ISO and Tanzanian standards incorporated with some modifications (ISO 9308-1,2000; Tzs 117,1981).
Study Site
The study was conducted along the Tanzanian basin of Lake Victoria in Mwanza. Sampling points included fishing grounds (open water), landing sites, and domestic fish markets located in Ilemela and Nyamagana districts in the Mwanza region. At the fishing grounds, there were no other activities besides fishing, and there were no toilet facilities on board. At some of the landing sites, there were households nearby and a range of different human activities including agriculture and scavenging animals, for example, cattle, goats, and dogs, around the landing sites. Also, there were several migratory fish-eating birds around the shore of the lake. At every landing site, there were toilet facilities, but it is uncertain whether these discharged into the lake. In the domestic markets, a variety of goods were sold, and toilets were present at all markets visited during sampling. However, there was poor waste management around the markets owing to accumulation and access of wastes in the garbage bins and also lack of proper wastewater discharge. Processing of samples and bacteriological analysis were done at the National Fish Quality Control Laboratory, Mwanza, and at the laboratory of the Department of Microbiology, Parasitology and Biotechnology, Sokoine University of Agriculture (SUA). The WGS of ESBL-producing E. coli was done at the Department of Veterinary and Animal Sciences, University of Copenhagen, Denmark.
Identification of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae
ESBL-producing Enterobacteriaceae in Nile perch and water were screened on MacConkey agar (Oxoid Ltd.) supplemented with 2 mg/L of cefotaxime (Moremi et al., 2016b). Briefly, 1 ml of an overnight enriched sample in BPW at 37°C was added to the MacConkey agar plates by the pour plate method and incubated overnight at 37°C (CLSI, 2016). Then, colonies with characteristics of Enterobacteriaceae were selected for ESBL confirmation using the double disk synergy method (Drieux et al., 2008; CLSI, 2016). The disks used were ceftazidime (30 μg) and cefotaxime (30 μg) (HiMedia Laboratories Pvt. Ltd., Mumbai, India), which were placed 20 mm apart from the center of a Mueller–Hinton agar (MHA) plate (Oxoid Ltd.) where amoxicillin/clavulanic acid (20 μg/10 μg) disk was placed. Plates were incubated at 37°C for 24 h. An inhibition zone around any of the disks under test increasing toward amoxicillin/clavulanic acid disk was interpreted as a presumptive ESBL-producing isolate, and the bacterial species were subsequently identified using API 20E (bioMérieux, France) based on the analytical profile index. The confirmed ESBL-producing E. coli isolates were further analyzed by PCR targeting β-lactamases genes – blaCTX–M, blaTEM, and blaSHV – in single multiplex reactions using primer sequences described by Nasreldin and Khaldoon (2015) to confirm their ESBL-producing status. The confirmed ESBL-producing isolates were used for further analyses.
Antimicrobial Disk Susceptibility Testing
The ESBL-producing E. coli isolates were subjected to antimicrobial susceptibility testing using standard disk diffusion method on MHA following the protocol and recommended antimicrobials by CLSI (2016). The following antimicrobials were selected: ampicillin/cloxacillin (AX; 10 μg), amoxicillin/clavulanic acid (AMC; 30 μg), gentamicin (GEN; 10 μg), sulfamethoxazole–trimethoprim (SXT; 25 μg), ciprofloxacin (CIP; 5 μg), cefotaxime (CTX; 30 μg), chloramphenicol (CHL; 30 μg), nalidixic acid (NAL; 30 μg), ceftazidime (CAZ; 30 μg), erythromycin (ERY; 15 μg), imipenem (IMP; 10 μg), and tetracycline (TET; 30 μg) (HiMedia Laboratories Pvt.). Although CLSI does not recommend the inclusion of erythromycin and ampicillin/cloxacillin, these antibiotics are commonly prescribed in Tanzania and were therefore included in the testing. E. coli ATCC 25922 was used as a quality control.
DNA Extraction and Whole Genome Sequencing
DNA was extracted from the 11 ESBL-producing E. coli isolates using Maxwell RSC culture cell’s DNA kit following the manufacturer’s protocol in the automated Maxwell RSC instrument (Promega, Fitchburg, WI, United States). Genomes were sequenced at the 250-bp paired-end-read format using Nextera XT kit and the MiSeq instrument (Illumina, Inc., San Diego, CA, United States). The average coverage of WGS and read length are indicated in Supplementary Table 3. Raw sequence reads have been submitted to the European Nucleotide Archive (ENA) under the project number PRJEB34642 with the accession numbers of each sample available in Supplementary Tables S1, S3 .
Data Analysis
Data collected were entered and stored into Microsoft Excel version 2010 (Microsoft Ltd., United States), and the prevalence of ESBL-producing E. coli in water and Nile perch at each sampling point was determined. E. coli genomes were de novo assembled using SPAdes 3.9.0 (Bankevich et al., 2012). Further analyses were performed using various web bioinformatics tools from the Center for Genomic Epidemiology (CGE),2 Enterobase,3 and BLASTn at National Centre for Biotechnology Information (NCBI).4 Plasmid replicons were detected using PlasmidFinder v2.0 (Carattoli et al., 2014), whereas virulence genes were determined using VirulenceFinder v.2.0 (Joensen et al., 2014), and In Silico analysis of resistance genes using ResFinder v2.2 (Zankari et al., 2012) from CGE using default settings.
Resistance to heavy metals and detergents was assessed using MyDbFinder 1.2 where our genomes were analyzed against plasmid-mediated genes such as the tellurite resistance gene tehA (NC_000913.3), the detergent-resistant phospholipase A, pldA (NC_003198.1), and the quaternary ammonium compound efflux qacEdelta (NG_048042.1) (Moremi et al., 2016b). Further heavy metal resistance operons encoding resistance to copper, cobalt, zinc, cadmium, magnesium, mercury, and chromium were analyzed through the subsystem annotation in RAST5 according to Brettin et al. (2015). The contigs harboring these genes and their positions on the contigs were compared with contigs harboring plasmids in the strains to confirm their location in the genomes. The location of β-lactamase genes on plasmids or chromosomes was determined by analyzing the contigs harboring the blaCTX–M–15 and blaTEM–1B using BLASTn in NCBI (Zhang et al., 2000).
Sequence types of the isolates were detected using In Silico MLST typing tool based on the seven housekeeping genes—adk, fumC, gyrB, icd, mdh, purA, and recA—for E. coli in Enterobase v.1.1.2 (Wirth et al., 2006). The phylogenetic analysis was performed using CSI Phylogeny v1.4 (Kaas et al., 2014) with the default options. The phylogenetic analysis included strains from this study and publicly available genomes of E. coli isolated from humans, foods, and the environments of the same sequence types as those identified for our strains. In addition to sequence type similarities, strains with the same serotype as our isolates were an added criterion in the selection of public genomes for phylogeny analysis. E. coli strain K12 sub-strain MG1655 (accession number SAMN02604091) was used as reference to root the tree. The list of E. coli strains used in phylogenetic analysis with their accession numbers is indicated in the table (Supplementary Table S1). The newick file of the tree was downloaded and edited using iTOL v46 (Letunic and Bork, 2016). The pairwise SNP difference data can be found in a table (Supplementary Table S2).
Results
Prevalence of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Nile Perch and Water
The overall prevalence of ESBL-producing Escherichia coli in Nile perch was 4.4% (8/180), whereas only one water sample collected offshore in the lake contained ESBL-producing E. coli. In this study, no other ESBL producers except for E. coli were detected. One Nile perch sample 1.7% (1/60) caught at fishing grounds offshore contained ESBL-producing E. coli, whereas two fish samples from landing sites contained ESBL-producing E. coli 3.3% (2/60) as did five fish samples purchased in domestic fish markets 8.3% (5/60) (Table 1).
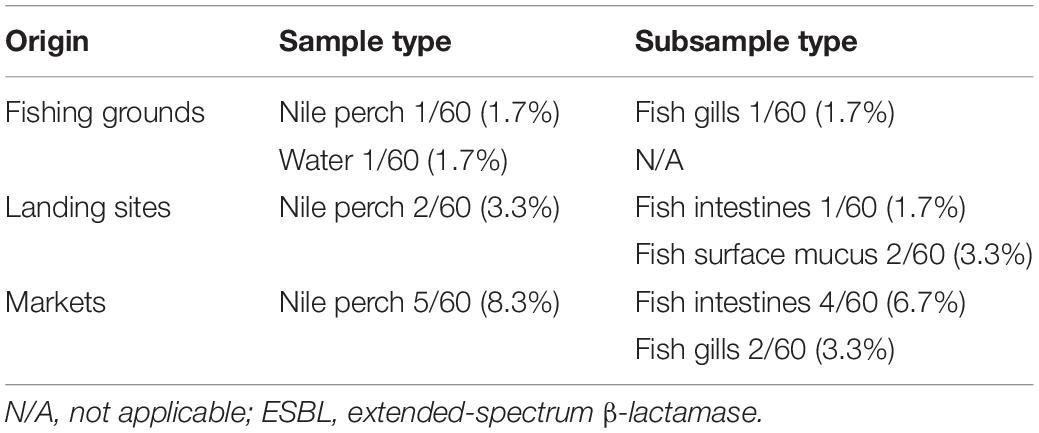
Table 1. Prevalence of ESBL-producing Escherichia coli in water and Nile perch from Lake Victoria, Tanzania.
Genomic Characteristics and Phylogenetic Analysis of Extended-Spectrum β-Lactamase-Producing Escherichia coli From Nile Perch
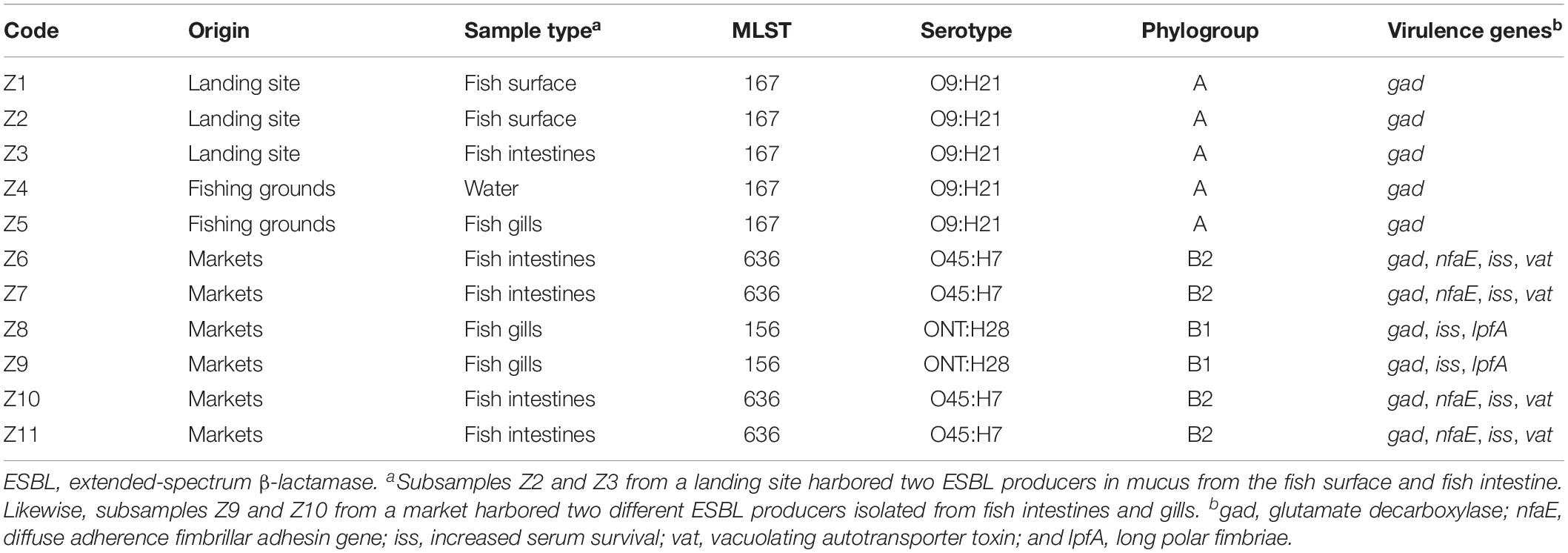
Analysis of the genome sequences from the 11 ESBL-producing E. coli revealed three clusters (C1, C2, and C3) separated by unique sequence type, serotype, phylogroup and also virulence and resistance genes patterns (Tables 2, 3). The cluster C1 included five clonal related strains (Z1 to Z5) with a SNP difference ranging from 3 to 13, all belonging to ST167, phylogroup A, and having serotype O9:H21. These strains possessed only one virulence gene (gad) and were isolated from fish and water samples collected at offshore fishing grounds, as well as from fish purchased at the landing sites. Subsample Z2 of mucus from the fish surface and Z3 (fish intestine) was from
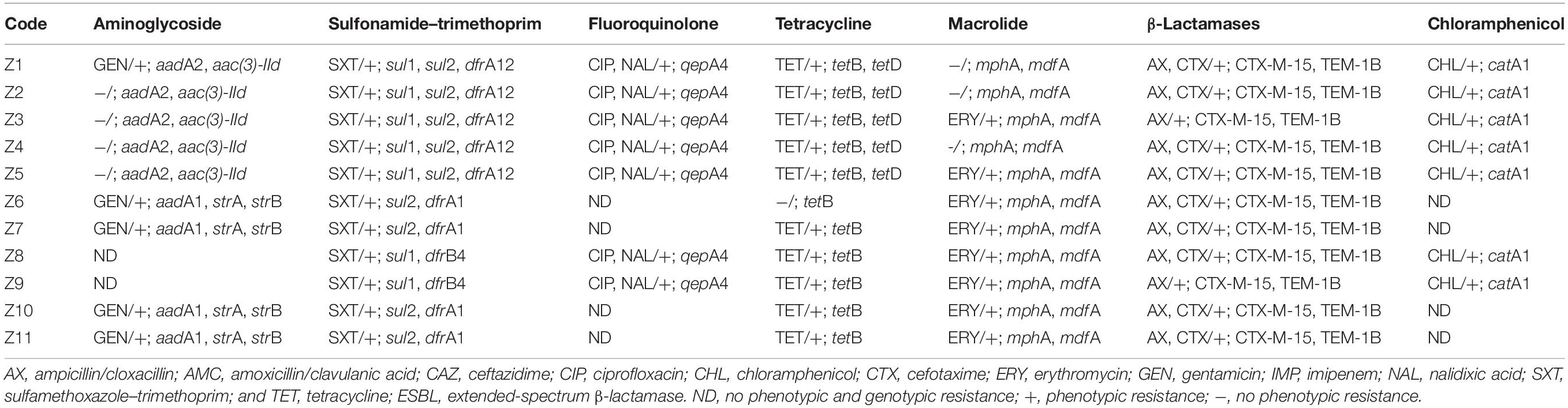
Table 3. Phenotypic and genotypic antimicrobial resistance of ESBL-producing Escherichia coli isolates.
the same fish sampled at a landing site, which harbored two ESBL producers. Likewise, subsamples Z9 and Z10 from the same fish collected at a market harbored two different ESBL producers isolated from fish intestines and gills. Two clonal strains (Z8 and Z9) formed cluster C2 with 11 SNPs apart. They belong to ST156, phylogroup B1, and showed serotype ONT:H28 and were both isolated from Nile perch obtained at local fish markets. These two strains contained additional virulence genes including gad, iss, lpfA, and nfaE.
Strains Z6, Z7, Z10, and Z11 isolated from Nile perch from fish markets formed the phylogenetic cluster C3 and had a SNP difference ranging from 4 to 17. These four strains belong to ST636, phylogroup B2; have serotype O45:H7; and harbor the vat gene in addition to all the virulence genes present in C2 strains. Among these isolates, we noted that one fish sample harbored two different ESBL producers (ST156 and ST636) in different sample subtypes, that is, from fish intestines and gills.
In a comparison with E. coli isolated from humans, animals, and the environment globally, our strains showed a sequence type-based clustering where the publically available ST167 genomes clustered with our C1 strains, as did the publically available ST156 and ST636 genomes with our C2 and C3 strains, respectively. However, we recorded some wide within-clade variations between our strains and the public genomes of the same STs with up to 5,104, 2,440, and 1,391 SNP differences in C1, C2, and C3, respectively, (Figure 1 and Supplementary Table S2).
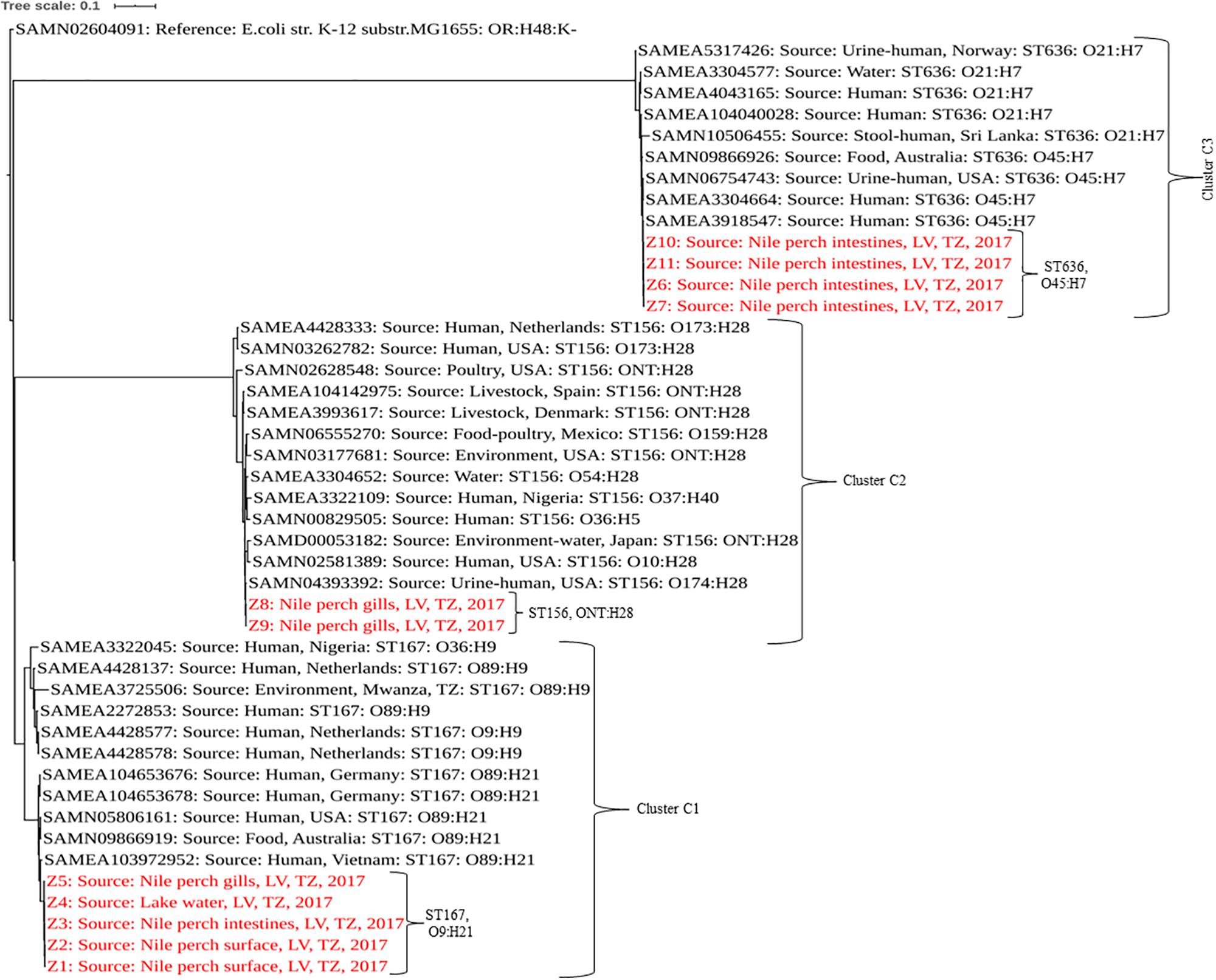
Figure 1. Phylogenetic tree based on SNPs showing clonal relationship among ESBL-producing E. coli isolated from Nile perch and global public genomes of E. coli strains. Our strains are highlighted in red color. LV, Lake Victoria; TZ, Tanzania.
Antimicrobial Resistance, Plasmid Replicon Profiles, Heavy Metal, and Detergent Resistance
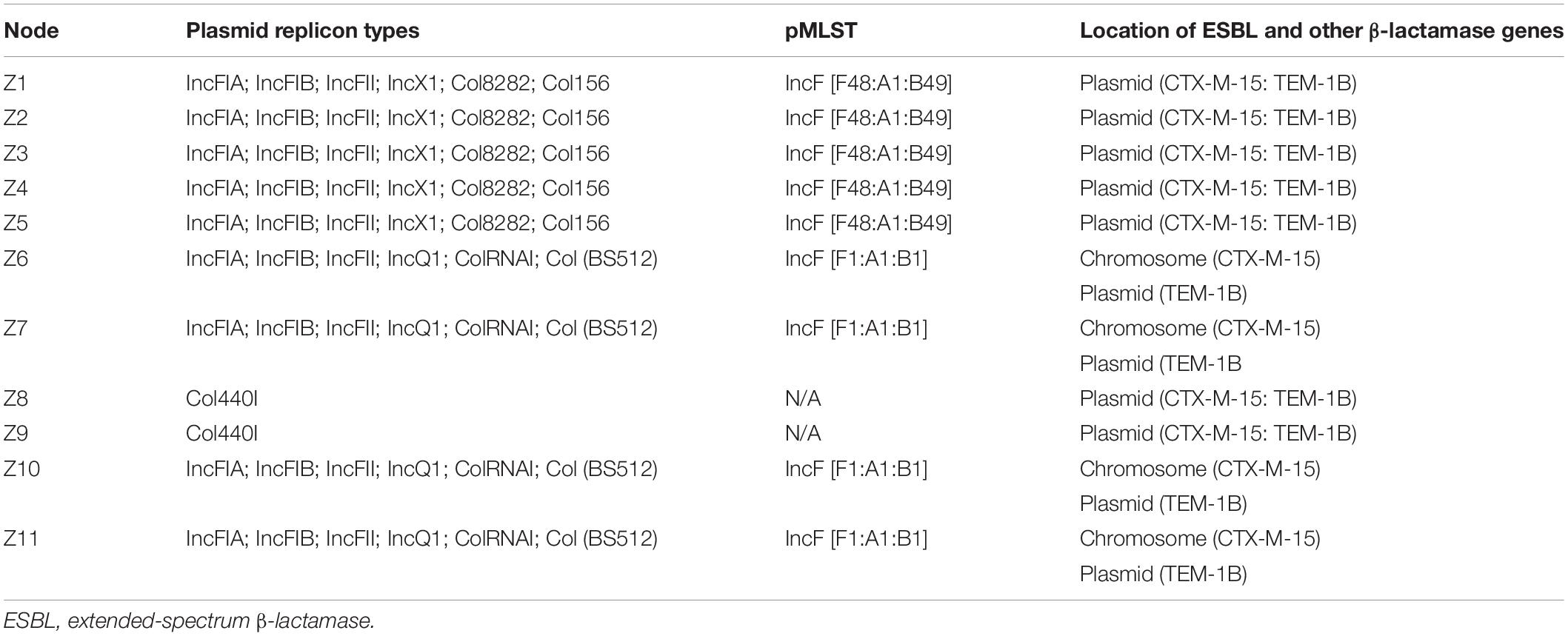
Overall, the genotype was consistent with the phenotypic resistance patterns, as well as occurrence of resistance genes and plasmid replicon types in the 11 ESBL-producing E. coli, which showed a similar clustering pattern as shown above (Tables 3, 4; Supplementary Table S3). The isolates showed phenotypic antimicrobial resistance to sulfamethoxazole–trimethoprim (100%), ampicillin/cloxacillin (100%), erythromycin (72.7%; 8/11), tetracycline (90.9%; 10/11), nalidixic acid (63.6%; 7/11), and chloramphenicol (63.6%; 7/11) (Table 3). C1 strains showed phenotypic and genotypic resistances to aminoglycosides [aadA2 and aac(3)-IId], sulfonamide–trimethoprim (sul1, sul2, and dfrA12), fluoroquinolone (qepA4), tetracycline (tetB and tetD), macrolides (mphA and mdfA), chloramphenicol (catA1), and the β-lactams ampicillin/cloxacillin and cefotaxime (blaCTX–M–15 and blaTEM–1B). However, strains Z1, Z2, and Z4 of C1 did not show phenotypic resistance to erythromycin despite the presence of mphA and mdfA conferring resistance to macrolides. All C1 strains have the plasmid replicon types IncFIA, IncFIB, IncFII, IncX1, Col8282, and Col156. Isolates contained IncF plasmids with the pMLST type [F48:A1:B49]. Strains Z8 and Z9 of clade C2 showed resistance to sulfonamide–trimethoprim (sul1 and dfrB4), fluoroquinolone (qepA4), tetracycline (tetB), macrolides (mphA and mdfA), chloramphenicol (catA1), and the β-lactams ampicillin/cloxacillin and cefotaxime (blaCTX–M–15 and blaTEM–1B). They were susceptible to aminoglycosides and only possessed one plasmid replicon type Col440I harboring the β-lactam genes.
The C3 strains were susceptible to fluoroquinolones and chloramphenicol but showed resistance to aminoglycosides (aadA1, strA, and strB), sulfonamide–trimethoprim (sul2 and dfrA1), tetracycline (tetB), macrolides (mphA and mdfA), and the β-lactams ampicillin/cloxacillin and cefotaxime (blaCTX–M–15 and blaTEM–1B). Despite the presence of tetB, strain Z6 did not show phenotypic resistance to tetracycline. All C3 strains have plasmid replicon types IncFIA, IncFIB, IncFII, IncQ1, ColRNAI, and Col (BS512) with the IncF harboring the blaTEM–1B gene, and they carried the blaCTX–M–15 on their chromosome. Isolates in this cluster contained IncF plasmids with the pMLST-type IncF [F1:A1:B1]. Strains in all three clusters were sensitive to imipenem.
All cluster C1 and C2 strains harbored the plasmid-mediated detergent resistance gene, qacEdelta, encoding resistance to quaternary ammonium compounds. All the strains contained resistance genes to copper, cobalt, zinc, cadmium, magnesium, mercury, and tellurite, which were chromosomally located (Supplementary Table S4). However, chromium resistance was only recorded in strains of cluster C1 isolated from fishing grounds and landing sites.
Discussion
The occurrence of ESBL-producing Escherichia coli was low in Nile perch and water samples collected at offshore fishing grounds and at landing sites of Lake Victoria but higher in fish from local fish markets in Mwanza. ESBL-producing E. coli have been reported in different wild-caught fish species in Switzerland, Algeria, and India (Abgottspon et al., 2014; Singh et al., 2017; Dib et al., 2018), as well as in farmed fish in China (Jiang et al., 2012) and Egypt (Ishida et al., 2010). Moremi et al. (2016b) reported a slightly higher prevalence of ESBL-producing bacteria in wild-caught tilapia sold at markets in Mwanza (13.3%) as compared with our findings in Nile perch (8.3%). Diverse bacterial species were found in the tilapia (Moremi et al., 2016b), whereas only E. coli was isolated in our Nile perch. This is probably because tilapia is usually found in shallow water with relatively high levels of fecal pollution and hence higher possibility of finding a variety of Enterobacteriaceae than finding these bacteria in the Nile perch, which are caught in deep water with lower pollution levels. Thus, these studies suggest that the aquatic environment and fish are likely sources and dissemination routes for ESBL-producing bacterial species and their resistance genes, although their relative importance from a public health perspective is uncertain.
The low prevalence of ESBL-producing E. coli in Nile perch from fishing grounds is likely due to the fact that these fish are commonly found and caught in deep water far away from the lake shore where fecal pollution is low compared with the shallow water near the shore of the lake (Baniga et al., 2019). Any occurrence of ESBL-producing E. coli in Nile perch caught offshore could be attributed to their eating habits feeding on other fish species, for example, tilapia and sardines, which are found in shallow water with higher fecal contamination levels. This hypothesis is corroborated by the findings of Moremi et al. (2016b) where gut intestinal samples from tilapia contained ESBL-producing bacteria. Because boats of artisanal small-scale fishers do not have toilet facilities, the fishermen have to defecate into the lake, with a potential introduction of ESBL-producing E. coli into the lake environment.
ESBL-producing E. coli isolated in Nile perch from markets belonged to clusters C2 and C3 and seem to harbor more virulence genes than do isolates originating from fish caught offshore. This higher pathogenicity indicates a human origin, and their occurrence in fish sold at markets is likely caused by fecal cross-contamination from fish handlers and vendors. This is further supported by the fact that most of the market strains of C3 belong to phylogroup B2, which is usually composed of extra-intestinal pathogenic E. coli strains (Abgottspon et al., 2014; Huang et al., 2016; Sánchez-Benito et al., 2017). It is important to note that our prevalence of ESBL-producing E. coli are generally very low, not allowing for any statistical comparison between fish of different origins. Moreover, possible associations between occurrence of ESBL-producing E. coli in fish and human handling activities can be better studied through comparative genomic investigations including clinical human isolates and our environmental strains.
The virulence gene gad harbored by C1 strains is an important gene for both commensal and pathogenic enteric bacteria especially E. coli, as the reaction products of gad are essential for survival in an acidic environment and for successful colonization of the host cell (Tramonti et al., 2002). The additional virulence genes shown by C2 and C3 strains included nfaE, which is essential for diffuse adherence fimbrillar adhesin; iss plays a significant role in increased serum survival; and the vat gene is important for vacuolating autotransporter toxin in host cells for pathogenicity processes. These virulence genes were previously reported from E. coli in humans as well as from pigs and bovine (Szmolka et al., 2012; Madoshi et al., 2016; Ahmed et al., 2017). The lpfA gene is important for long polar fimbriae and has been commonly associated with the ability to invade epithelial cells in animals and humans (Dogan et al., 2012).
Our strains were epidemiologically unrelated, but their grouping into three clusters with further clustering with other E. coli strains of the same STs isolated from human, food, and environmental samples around the globe underlines the transmission potential of the ESBL genes across various niches and locations favored by plasmids that they harbor. Our strains of cluster C1 (ST167) belong to the phylogroup A known to contain commensal strains (Abgottspon et al., 2014; Huang et al., 2016; Sánchez-Benito et al., 2017), and this is supported by the presence of only one virulence gene (gad). Moremi et al. (2016b) reported a single environmental sample from Mwanza containing an ESBL-producing E. coli of ST167, belonging to the newly defined phylogroup E harboring blaCTX–M–15 and blaOXA–1. This ST167 strain of phylogroup E differed from our strains of ST167 of phylogroup A with up to 5,104 SNPs. The variation among strains of the same STs can be attributed to the differences in serotype as well as their genetic content such as the differences in the ESBL genes that they contained. Our study corroborates earlier findings that blaCTX–M–15 is the predominant ESBL gene in Enterobacteriaceae in Tanzania (Moremi et al., 2016b), yet the sources are still unknown. ESBL-producing E. coli ST167 have been previously reported isolated in humans in China and Spain carrying blaCTX–M–15 and blaTEM–1 genes (Huang et al., 2016; Sánchez-Benito et al., 2017). This substantiates the possibilities of horizontal gene transfer of the ESBL genes from food products or the environment to humans where bacterial pathogens can acquire them. Such transmission is favored by plasmids on which the resistance genes are located (Anjum et al., 2019) as is the case for the bla-CTXM–15 and blaTEM–1 genes, which were located on the IncF plasmid (Table 4). We observed the plasmid IncF [F1:A1:B1] within all four C3 strains, which had the same ST636 and the same pMLST and showed a SNP difference ranging from 4 to 17. The C3 strains all came from intestinal samples, and we cannot explain their clonal nature as the origin and previous exposures of the fish purchased at the markets are unknown. The IncF [F48:A1:B49] was shown in all five C1 strains originating from intestinal and surface mucus samples of fish from one landing site as well as from fish (gills) and water collected offshore. Similarly to C3 strains, the C1 strains were epidemiologically unrelated, and we are not able to explain the clonal nature of the C1 strains. In contrast to our study, the ESBL-producing bacteria reported in tilapia from the Mwanza region by Moremi et al. (2016b) included a much more diverse population of bacterial species, a variety of ESBL genes, and no indication of a clonal relationship. The occurrence of genes encoding for resistance to metals such as copper, cobalt, zinc, cadmium, and mercury, and also genes encoding resistance to detergents is similar to previous findings in fish and aquatic environment of the Lake Victoria; the genes have been reported to play a role in environmental persistence support in bacterial survival (Moremi et al., 2016b; Hounmanou et al., 2019). Most of metal resistance genes were located on the chromosome, whereas ESBL genes were mostly located on the plasmids, except that all four ST636 isolates harbored a chromosomally located blaCTX–M–15 gene, and it is uncertain to what extent exposure to metals, for example, used as livestock feed additives and as pollutants in aquatic environments, may play a role as co-selectors of ESBL resistance.
In addition to the ESBL genes, our strains are all multidrug resistant despite their relatively low virulence (Magiorakos et al., 2012). The resistance genes found have been frequently reported in Gram-negative bacteria, and because the genes are often plasmid mediated, they may be circulated horizontally among different bacterial species (van Hoek et al., 2011). These additional resistances shown by the ESBL-producing E. coli may also reflect the frequent use of antimicrobials reported in human and veterinary medicine in the Mwanza region (Moremi et al., 2016a; Seni et al., 2016). The location of the additional resistance genes requires further analysis and depiction of the genetic environment of blaCTX–M15 genes in the blaCTX–M–15-encoding isolates (Moremi et al., 2016b).
Some strains in C1 and C3 did not show phenotypic resistance to antimicrobials that they harbored resistance genes for, that is, erythromycin and tetracycline. This is an increasing observation in genomic studies and could be due to various factors including the concentration and quality of the antimicrobial disks (Eze et al., 2014) or intrinsic factors inhibiting the expression of the resistance genes (Weill et al., 2017). This has also been associated with random mutations, which could be accumulated in gene sequences encoding resistance to some antimicrobials (Davis et al., 2011; Hussain et al., 2014).
Conclusion
In conclusion, we report a very low prevalence of ESBL-producing E. coli in Nile perch from Lake Victoria. Our data suggest that as far as ESBL-producing enteric bacteria are concerned, the consumption of Nile perch represent limited food safety risks compared with other human exposures to ESBL-producing E. coli, for example, through direct human-to-human fecal transmission and consumption of livestock meat products. The grouping of the 11 ESBL-producing E. coli into three clades each showing identical characteristics, for example, STs, phylogroup, antimicrobial resistance, and virulence genes, is surprising, and we are not able to explain the clonal nature of these clades as the E. coli strains were epidemiologically unrelated. All isolates harbored blaCTX–M–15 and blaTEM–1 genes together with additional antimicrobial and detergent resistance genes carried by the common plasmids replicon types IncF, IncX, IncQ, and Col. Further studies are needed to determine the role of fish and aquatic environments as sources of ESBL-producing bacteria and resistance genes including the importance of fecal pollution sources, for example, discharge of sewage and runoff water from fertilized agricultural soil, as well as the ecology of such resistant bacteria in aquatic environments.
Our study has some limitations, including failure to obtain other Enterobacteriaceae spp. than E. coli. This is likely because of the selective isolation procedure used with supplement of cefotaxime to obtain ESBL-producing strains; thus, most of other bacteria present both in deep water and in fish from markets were likely sensitive to cefotaxime and did therefore not grow on the MacConkey agar. Although E. coli isolated from Nile perch at fish markets were of different sequence types and harbored more virulence genes than did isolates from deep water, a comparison study with ESBL-producing E. coli from clinical specimens with environmental strains may provide further information about transmission with the aquatic environment, fish, and human compartments.
Data Availability Statement
Raw sequence reads have been submitted to the European Nucleotide Archive (ENA) under the project number PRJEB34642.
Ethics Statement
The present study required no ethical approval because we did not manipulate any live fish in the study. Fish samples used in the study were fished by fishermen and constitute dead but fresh fish products ready for marketing and consumption. We purchased these fish from the fishermen, in boats at landing sites and in the markets from fish retailers as indicated in the manuscript like any other fish consumer, and then we brought them to the laboratory for analyses.
Author Contributions
ZB collected samples, carried out laboratory and data analysis, and wrote the draft manuscript. YH participated in laboratory analysis and performed genomic data analysis and editing and critical review of the manuscript. EK participated in genomic data analysis and reviewed the manuscript. LK and RM supervised the study and were involved in reviewing the manuscript and facilitation of resources for the study. AD supervised the study and provided guidance and resources, contributed in genomic data analysis, revised the manuscript, and gave the final approval of the manuscript. All authors read and approved the final manuscript for submission.
Funding
This study was funded by the “Innovations and Markets for Lake Victoria Fisheries” (IMLAF) project, under the Danish International Development Agency (DANIDA) project code number IMLAF, DFC file no. 14-P01-TAN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the staff at the National Fish Quality Control Laboratory in Mwanza; the Department of Microbiology, Parasitology and Biotechnology at SUA; and the Department of Veterinary and Animal Sciences at the University of Copenhagen for their technical assistance offered during sample analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00108/full#supplementary-material
Footnotes
- ^ www.seward.co.uk
- ^ https://cge.cbs.dtu.dk/services/
- ^ http://enterobase.warwick.ac.uk/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://rast.nmpdr.org/rast.cgi
- ^ https://itol.embl.de/
References
Abgottspon, H., Nüesch-Inderbinen, M. T., Zurfluh, K., Althaus, D., Hächler, H., and Stephan, R. (2014). Enterobacteriaceae with extended-spectrum- and pAmpC-type β-lactamase-encoding genes isolated from freshwater fish from two lakes in Switzerland. Antimicrob. Agents Chemother. 58, 2482–2484. doi: 10.1128/AAC.02689-13
Adelowo, O. O., Caucci, S., Banjo, O. A., Nnanna, O. C., Awotipe, E. O., Peters, F. B., et al. (2018). Extended spectrum beta-lactamase (ESBL)-producing bacteria isolated from hospital wastewaters, rivers and aquaculture sources in Nigeria. Environ. Sci. Pollut. Res. 25, 2744–2755. doi: 10.1007/s11356-017-0686-7
Ahmed, S., Olsen, J. E., and Herrero-Fresno, A. (2017). The genetic diversity of commensal Escherichia coli strains isolated from non-antimicrobial treated pigs varies according to age group. PLoS One 12:e0178623. doi: 10.1371/journal.pone.0178623
Anjum, M., Madsen, J. S., Nesme, J., Jana, B., Wiese, M., Jasinskytė, D., et al. (2019). Fate of CMY-2-Encoding plasmids introduced into the human fecal microbiota by exogenous Escherichia coli. Antimicrob. Agents Chemother 63:e02528. doi: 10.1128/AAC.02528-18
Baniga, Z., Mdegela, R. H., Lisa, B., Kusiluka, L. J. M., and Dalsgaard, A. (2019). Prevalence and characterisation of Salmonella Waycross and Salmonella enterica subsp. salamae in Nile perch (Lates niloticus) of Lake Victoria. Tanzania. Food Control 100, 28–34. doi: 10.1016/j.foodcont.2019.01.006
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Ben Said, L., Jouini, A., Klibi, N., Dziri, R., Alonso, C. A., Boudabous, A., et al. (2015). Detection of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. Int. J. Food Microbiol. 203, 86–92. doi: 10.1016/j.ijfoodmicro.2015.02.023
Blaak, H., de Kruijf, P., Hamidjaja, R. A., van Hoek, A. H., de Roda Husman, A. M., and Schets, F. M. (2014). Prevalence and characteristics of ESBL-producing E. coli in Dutch recreational waters influenced by wastewater treatment plants. Vet. Microbiol. 171, 448–459. doi: 10.1016/j.vetmic.2014.03.007
Boonyasiri, A., Tangkoskul, T., Seenama, C., Saiyarin, J., Tiengrim, S., and Thamlikitkul, V. (2014). Prevalence of antibiotic resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathog. Glob. Health 108, 235–245. doi: 10.1179/2047773214Y.0000000148
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In Silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chishimba, K., Hang’ombe, B. M., Muzandu, K., Mshana, S. E., Matee, M. I., Nakajima, C., et al. (2016). Detection of extended-spectrum beta-lactamase-producing Escherichia coli in market-ready chickens in Zambia. Int. J. Microbiol. 2016, 1–5. doi: 10.1155/2016/5275724
CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing, 26th Edn. CLSI Supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute.
Davis, M. A., Besser, T. E., Orfe, L. H., Baker, K. N. K., Lanier, A. S., Broschat, S. L., et al. (2011). Genotypic-phenotypic discrepancies between antibiotic resistance characteristics of Escherichia coli isolates from calves in management settings with high and low antibiotic use. Appl. Environ. Microbiol. 77, 3293–3299. doi: 10.1128/AEM.02588-10
Delgado, D. Y. C., Barrigas, Z. P. T., Astutillo, S. G. O., Jaramillo, A. P. A., and Ausili, A. (2016). Detection and molecular characterization of β-lactamase genes in clinical isolates of Gram-negative bacteria in Southern Ecuador. Braz. J. Infect. Dis. 20, 627–630. doi: 10.1016/j.bjid.2016.07.001
Dib, A. L., Agabou, A., Chahed, A., Kurekci, C., Moreno, E., Espigares, M., et al. (2018). Isolation, molecular characterization and antimicrobial resistance of Enterobacteriaceae isolated from fish and seafood. Food Control 88, 54–60. doi: 10.1016/j.foodcont.2018.01.005
Dogan, B., Rishniw, M., Bruant, G., Harel, J., Schukken, Y. H., and Simpson, K. W. (2012). Phylogroup and lpfA influence epithelial invasion by mastitis associated Escherichia coli. Vet. Microbiol. 159, 163–170. doi: 10.1016/j.vetmic.2012.03.033
Drieux, L., Brossier, F., Sougakoff, W., and Jarlier, V. (2008). Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin. Microbiol. Infect. 14, 90–103. doi: 10.1111/j.1469-0691.2007.01846.x
Egea, P., López-Cerero, L., Torres, E., Gómez-Sánchez, M., del, C., Serrano, L., et al. (2012). Increased raw poultry meat colonization by extended spectrum beta-lactamase-producing Escherichia coli in the south of Spain. Int. J. Food Microbiol. 159, 69–73. doi: 10.1016/j.ijfoodmicro.2012.08.002
Eze, P., Ajaegbu, E., Ejikeugwu, P., Egbuna, R., Abba, C., and Esimone, C. (2014). Evaluation of the quality of commercial antibacterial discs available in Nigeria. Br. J. Pharm. Res. 4, 2548–2562. doi: 10.9734/BJPR/2014/10900
Falgenhauer, L., Imirzalioglu, C., Oppong, K., Akenten, C. W., Hogan, B., Krumkamp, R., et al. (2019). Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front. Microbiol. 9:3358. doi: 10.3389/fmicb.2018.03358
Hounmanou, Y. M. G., Leekitcharoenphon, P., Hendriksen, R. S., Dougnon, T. V., Mdegela, R. H., Olsen, J. E., et al. (2019). Surveillance and genomics of toxigenic Vibrio cholerae O1 from fish, phytoplankton and water in Lake Victoria. Tanzania. Front. Microbiol. 10:901. doi: 10.3389/fmicb.2019.00901
Huang, Y., Yu, X., Xie, M., Wang, X., Liao, K., Xue, W., et al. (2016). Widespread Dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring bla NDM-5 in clinical settings in China. Antimicrob. Agents Chemother. 60, 4364–4368. doi: 10.1128/AAC.00859-16
Hussain, A., Ranjan, A., Nandanwar, N., Babbar, A., Jadhav, S., and Ahmed, N. (2014). Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob. Agents Chemother. 58, 7240–7249. doi: 10.1128/AAC.03320-14
Ishida, Y., Ahmed, A. M., Mahfouz, N. B., Kimura, T., El-Khodery, S. A., Moawad, A. A., et al. (2010). Molecular analysis of antimicrobial resistance in Gram-negative bacteria isolated from fish farms in Egypt. J. Vet. Med. Sci. 72, 727–734. doi: 10.1292/jvms.09-0538
ISO 6887-3 (2003). Microbiology of Food and Animal Feeding Stuffs — Preparation of Test Samples, Initial Suspension and Decimal Dilutions for Microbiological Examination — Part 3: Specific Rules for the Preparation of Fish and Fishery Products. Geneva: ISO. doi: 10.1292/jvms.09-0538
ISO 9308-1 (2000). Water Quality — Detection and Enumeration of Escherichia coli and Coliform Bacteria — Part 1: Membrane Filtration Method. Geneva: ISO. doi: 10.1292/jvms.09-0538
Jiang, H.-X., Tang, D., Liu, Y.-H., Zhang, X.-H., Zeng, Z.-L., Xu, L., et al. (2012). Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 67, 2350–2353. doi: 10.1093/jac/dks250
Jiang, L., Hu, X., Xu, T., Zhang, H., Sheng, D., and Yin, D. (2013). Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu river and the drinking water sources. Shanghai, China. Sci. Total Environ. 45, 267–272. doi: 10.1016/j.scitotenv.2013.04.038
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Katakweba, A. A. S., Muhairwa, A. P., Lupindu, A. M., Damborg, P., Rosenkrantz, J. T., Minga, U. M., et al. (2018). First report on a randomized investigation of antimicrobial resistance in fecal indicator bacteria from livestock, poultry, and humans in Tanzania. Microb. Drug Resist. 24, 260–268. doi: 10.1089/mdr.2016.0297
Kümmerer, K. (2009). Antibiotics in the aquatic environment – A review – Part I. Chemosphere 75, 417–434. doi: 10.1016/j.chemosphere.2008.11.086
Lavilla, S., Gonzalez-Lopez, J. J., Miro, E., Dominguez, A., Llagostera, M., Bartolome, R. M., et al. (2008). Dissemination of extended-spectrum β-lactamase-producing bacteria: the food-borne outbreak lesson. J. Antimicrob. Chemother. 61, 1244–1251. doi: 10.1093/jac/dkn093
Legese, M. H., Weldearegay, G. M., and Asrat, D. (2017). Extended-spectrum beta-lactamase and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect. Drug Resist. Volume 10, 27–34. doi: 10.2147/IDR.S127177
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Madoshi, B. P., Kudirkiene, E., Mtambo, M. M. A., Muhairwa, A. P., Lupindu, A. M., and Olsen, J. E. (2016). Characterisation of commensal Escherichia coli isolated from apparently healthy cattle and their attendants in Tanzania. PloS One 11:e0168160. doi: 10.1371/journal.pone.0168160
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Martinez, J. L. (2009). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. B Biol. Sci. 276, 2521–2530. doi: 10.1098/rspb.2009.0320
Moremi, N., Claus, H., and Mshana, S. E. (2016a). Antimicrobial resistance pattern: a report of microbiological cultures at a tertiary hospital in Tanzania. BMC Infect. Dis. 16:756. doi: 10.1186/s12879-016-2082-1
Moremi, N., Manda, E. V., Falgenhauer, L., Ghosh, H., Imirzalioglu, C., Matee, M., et al. (2016b). Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front. Microbiol 7:1862. doi: 10.3389/fmicb.2016.01862
Moyo, S. J., Aboud, S., Kasubi, M., Lyamuya, E. F., and Maselle, S. Y. (2010). Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC Res. Notes 3:348. doi: 10.1186/1756-0500-3-348
Nasreldin, E., and Khaldoon, A. (2015). Incidence and antimicrobial susceptibility pattern of extended-spectrum–lactamase-producing Escherichia coli isolated from retail imported mackerel fish. Afr. J. Biotechnol. 14, 1954–1960. doi: 10.5897/AJB2015.14698
Nguyen, D. T. A., Kanki, M., Nguyen, P. D., Le, H. T., Ngo, P. T., Tran, D. N. M., et al. (2016). Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City. Vietnam. Int. J. Food Microbiol. 236, 115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017
Ojer-Usoz, E., González, D., Vitas, A. I., Leiva, J., García-Jalón, I., Febles-Casquero, A., et al. (2013). Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in meat products sold in Navarra. Spain. Meat Sci. 93, 316–321. doi: 10.1016/j.meatsci.2012.09.009
Overdevest, I. (2011). Extended-Spectrum B-Lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg. Infect. Dis. 17, 1216–1222. doi: 10.3201/eid1707.110209
Projahn, M., von Tippelskirch, P., Semmler, T., Guenther, S., Alter, T., and Roesler, U. (2019). Contamination of chicken meat with extended-spectrum beta-lactamase producing Klebsiella pneumoniae and Escherichia coli during scalding and defeathering of broiler carcasses. Food Microbiol. 77, 185–191. doi: 10.1016/j.fm.2018.09.010
Rupp, M. E., and Fey, P. D. (2003). Extended spectrum β-Lactamase (ESBL)-producing Enterobacteriaceae: considerations for diagnosis, prevention and drug treatment. Drugs 63, 353–365. doi: 10.2165/00003495-200363040-00002
Sánchez-Benito, R., Iglesias, M. R., Quijada, N. M., Campos, M. J., Ugarte-Ruiz, M., Hernández, M., et al. (2017). Escherichia coli ST167 carrying plasmid mobilisable mcr-1 and bla CTX-M-15 resistance determinants isolated from a human respiratory infection. Int. J. Antimicrob. Agents 50, 285–286. doi: 10.1016/j.ijantimicag.2017.05.005
Seni, J., Falgenhauer, L., Simeo, N., Mirambo, M. M., Imirzalioglu, C., Matee, M., et al. (2016). Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front. Microbiol 7:142. doi: 10.3389/fmicb.2016.00142
Shaikh, S., Fatima, J., Shakil, S., Rizvi, S., Mohd, D., and Kamal, M. A. (2015). Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J. Biol. Sci. 22, 90–101. doi: 10.1016/j.sjbs.2014.08.002
Singh, A. S., Lekshmi, M., Prakasan, S., Nayak, B. B., and Kumar, S. (2017). Multiple antibiotic-resistant, extended spectrum-β-Lactamase (ESBL)-producing Enterobacteria in fresh seafood. Microorganisms 5:53. doi: 10.3390/microorganisms5030053
Smet, A., Martel, A., Persoons, D., Dewulf, J., Heyndrickx, M., Herman, L., et al. (2010). Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 34, 295–316. doi: 10.1111/j.1574-6976.2009.00198.x
Szmolka, A., Anjum, M. F., La Ragione, R. M., Kaszanyitzky, ÉJ., and Nagy, B. (2012). Microarray based comparative genotyping of gentamicin resistant Escherichia coli strains from food animals and humans. Vet. Microbiol. 156, 110–118. doi: 10.1016/j.vetmic.2011.09.030
Tramonti, A., Visca, P., De Canio, M., Falconi, M., and De Biase, D. (2002). Functional characterization and regulation of gadX, a gene encoding an araC/xylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184, 2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002
Tzs 117. (1981). Tanzania Tanzania Standard: Food-Handling of Samples for Microbiological Analysis UDC 664:579.67.086. Dar es Salaam.
van Hoek, A. H. A. M., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., and Aarts, H. J. M. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2:203. doi: 10.3389/fmicb.2011.00203
Weill, F.-X., Domman, D., Njamkepo, E., Tarr, C., Rauzier, J., Fawal, N., et al. (2017). Genomic history of the seventh pandemic of cholera in Africa. Science 358, 785–789. doi: 10.1126/science.aad5901
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: nile perch, food safety, extended-spectrum β-lactamase, antimicrobial resistance, Escherichia coli
Citation: Baniga Z, Hounmanou YMG, Kudirkiene E, Kusiluka LJM, Mdegela RH and Dalsgaard A (2020) Genome-Based Analysis of Extended-Spectrum β-Lactamase-Producing Escherichia coli in the Aquatic Environment and Nile Perch (Lates niloticus) of Lake Victoria, Tanzania. Front. Microbiol. 11:108. doi: 10.3389/fmicb.2020.00108
Received: 13 August 2019; Accepted: 17 January 2020;
Published: 21 February 2020.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Stephen E. Mshana, Catholic University of Health and Allied Sciences (CUHAS), TanzaniaLinda Falgenhauer, University of Giessen, Germany
Copyright © 2020 Baniga, Hounmanou, Kudirkiene, Kusiluka, Mdegela and Dalsgaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zebedayo Baniga, zebe_02@yahoo.co.uk; Yaovi M. Gildas Hounmanou, gil@sund.ku.dk
 Zebedayo Baniga
Zebedayo Baniga Yaovi M. Gildas Hounmanou
Yaovi M. Gildas Hounmanou Egle Kudirkiene
Egle Kudirkiene Lughano J. M. Kusiluka1,4
Lughano J. M. Kusiluka1,4 Anders Dalsgaard
Anders Dalsgaard