- 1bioMérieux, Data Analytics Unit, La Balme-les-Grottes, France
- 2Department of Biology, University of “Tor Vergata”, Rome, Italy
- 3Department of Medical Biotechnologies, University of Siena, Siena, Italy
- 4bioMérieux, R&D Microbiology, La Balme-les-Grottes, France
- 5Institute for Laboratory Diagnostics Konzilijum, Belgrade, Serbia
- 6Institute of Microbiology and Immunology, Faculty of Medicine, University of Belgrade, Belgrade, Serbia
- 7Laboratory of Medical Microbiology, Vaccine and Infectious Disease Institute, University of Antwerp, Antwerp, Belgium
- 8Microbiology and Virology Unit, Florence Careggi University Hospital, Florence, Italy
- 9Department of Experimental and Clinical Medicine, University of Florence, Florence, Italy
Klebsiella pneumoniae is a major cause of severe healthcare-associated infections and often shows MDR phenotypes. Carbapenem resistance is frequent, and colistin represents a key molecule to treat infections caused by such isolates. Here we evaluated the antimicrobial resistance (AMR) mechanisms and the genomic epidemiology of clinical K. pneumoniae isolates from Serbia. Consecutive non-replicate K. pneumoniae clinical isolates (n = 2,298) were collected from seven hospitals located in five Serbian cities and tested for carbapenem resistance by disk diffusion. Isolates resistant to at least one carbapenem (n = 426) were further tested for colistin resistance with Etest or Vitek2. Broth microdilution (BMD) was performed to confirm the colistin resistance phenotype, and colistin-resistant isolates (N = 45, 10.6%) were characterized by Vitek2 and whole genome sequencing. Three different clonal groups (CGs) were observed: CG101 (ST101, N = 38), CG258 (ST437, N = 4; ST340, N = 1; ST258, N = 1) and CG17 (ST336, N = 1). mcr genes, encoding for acquired colistin resistance, were not observed, while all the genomes presented mutations previously associated with colistin resistance. In particular, all strains had a mutated MgrB, with MgrBC28S being the prevalent mutation and associated with ST101. Isolates belonging to ST101 harbored the carbapenemase OXA-48, which is generally encoded by an IncL/M plasmid that was no detected in our isolates. MinION sequencing was performed on a representative ST101 strain, and the obtained long reads were assembled together with the Illumina high quality reads to decipher the blaOXA–48 genetic background. The blaOXA–48 gene was located in a novel IncFIA-IncR hybrid plasmid, also containing the extended spectrum β-lactamase-encoding gene blaCTX–M–15 and several other AMR genes. Non-ST101 isolates presented different MgrB alterations (C28S, C28Y, K2∗, K3∗, Q30∗, adenine deletion leading to frameshift and premature termination, IS5-mediated inactivation) and expressed different carbapenemases: OXA-48 (ST437 and ST336), NDM-1 (ST437 and ST340) and KPC-2 (ST258). Our study reports the clonal expansion of the newly emerging ST101 clone in Serbia. This high-risk clone appears adept at acquiring resistance, and efforts should be made to contain the spread of such clone.
Introduction
Klebsiella pneumoniae has emerged as one of the most challenging antibiotic-resistant pathogens, since it can cause a variety of infections, including pneumonia and bloodstream infections, and exhibits a remarkable propensity to acquire antimicrobial resistance (AMR) traits. In particular, carbapenem-resistant K. pneumoniae (CRKP) are challenging pathogens due to the limited treatment options, high mortality rates, and potential for rapid dissemination in health care settings (Paczosa and Mecsas, 2016).
Treatment options for CRKP infections are usually limited to aminoglycosides, tigecycline, fosfomycin, and colistin. Novel β-lactam-β-lactamase inhibitors combinations, such as ceftazidime-avibactam and meropenem-vaborbactam, have represented a major breakthrough for treatment of some CRKP (e.g., those producing KPC-type and OXA-48-like enzymes), but unfortunately they do not cover strains producing metallo-carbapenemases (Bassetti et al., 2018). Colistin, despite its nephrotoxicity and neurotoxicity, remains a key component of some anti-CRKP regimens (Karaiskos et al., 2017).
Colistin resistance (colR) is mainly mediated by modifications of the lipid A moiety of the bacterial lipopolysaccharide (LPS) by addition of positively charged 4-amino-4-deoxy-L-arabinose (LAra4N) and/or phosphoethanolamine (pEtN) residues. A large panel of genes and operons is involved in modifications of the LPS, and mutations conferring colistin resistance have mainly been observed in mgrB, phoP/phoQ, pmrA/pmrB, and crrB genes (Cheng et al., 2010; Cannatelli et al., 2013, 2014a; Wright et al., 2015). Recently, several plasmid-mediated colistin resistance genes, named mcr, encoding pEtN transferases, have also been reported in E. coli and other members of Enterobacterales, including K. pneumoniae (Sun et al., 2018).
Global dissemination of CRKP is mainly caused by the spread of a few successful clones. Major representatives of these high-risk clonal lineages include the clonal group (CG) 11, CG15, CG307, CG17, CG37, CG101, and CG147 strains. CG258 strains, and in particular those of ST258, are major players in the worldwide spread of KPC-type carbapenemases, and are responsible for 68% of the CRKP outbreaks (Navon-Venezia et al., 2017). CG101 strains harbor different clinically-relevant resistance determinants, such as carbapenemases of the KPC, OXA-48, VIM, and NDM types. This feature, together with their ability to produce biofilm and several additional virulence factors, is likely a major factor in the ecological success of CG101 strains. Indeed, spreading of this clone is on the rise (Navon-Venezia et al., 2017).
Multidrug resistance (MDR) prevalence in clinical isolates of K. pneumoniae, including resistance to third-generation cephalosporins, fluoroquinolones and aminoglycosides, may be as high as 50% in Southern Europe, and even higher proportions have been observed in Eastern Europe. In Serbia, in 2016, MDR K. pneumoniae accounted for 63% of all K. pneumoniae infections in humans, of which 35% were also carbapenem resistant (WHO Regional Office for Europe, 2017). Previous studies reported that NDM-1 was the main K. pneumoniae-associated carbapenemase observed in Serbia in the period 2013–2014 followed by OXA-48, while KPC was only sporadically reported (Grundmann et al., 2017; Trudic et al., 2017). Novović et al. (2017) performed a molecular epidemiology study of carbapenem- and colistin-resistant strains from Serbia, showing prevalence of CG258 and CG101 strains, producing NDM-1 and OXA-48 carbapenemases, respectively. However, the proportion of colistin resistance among those isolates was not reported, and the mechanisms of colistin resistance of those isolates were not elucidated (Novović et al., 2017).
In this study, we used whole genome sequencing (WGS) to study the genomic epidemiology and AMR mechanisms of colR K. pneumoniae isolates from Serbia, including some representative of the previously mentioned collection as reference to study the dynamic changes of population structure (Novović et al., 2017).
Materials and Methods
Bacterial Isolates and Susceptibility Testing
In the period between November 2013 and May 2017, K. pneumoniae isolates were obtained from routine microbiological cultures of clinical samples (e.g., urine, blood, skin, bronchial aspirate) from seven Serbian medical centers distributed in five Serbian cities (Niš, Novi Sad, Belgrade, Kraljevo, and Subotica). Bacteria were not isolated by the authors but provided by the respective medical centers. Therefore, an ethics approval was not required as per institutional and national guidelines and regulations. Information about patients antimicrobial treatment were not available. Identification at the species level was performed by Vitek2 (bioMérieux, Marcy-l’Etoile, France), and carbapenem susceptibility was determined by disk diffusion and interpreted according to the EUCAST breakpoints (EUCAST, 2019). Isolates non-susceptible to at least one carbapenem (ertapenem, meropenem, and imipenem) were tested for colistin resistance by Vitek2 or Etest (bioMérieux, Marcy-l’Etoile, France) according to manufacturer’s instructions (note that the warning by EUCAST about colistin susceptibility testing was only issued in July 2016, and for this reason the above methods were used for colistin susceptibility testing of the isolates collected in this study). Antimicrobial susceptibility testing of the colR isolates was performed using the Vitek2 automated system, and results were interpreted according to EUCAST breakpoints (EUCAST, 2019). Colistin minimum inhibitory concentrations (MICs) were confirmed using the broth microdilution method performed according to the CLSI guidelines (CLSI, 2019) and interpreted by using the EUCAST breakpoints (EUCAST, 2019). For carbapenems (ertapenem, imipenem, and meropenem), MICs were obtained by using Etests (bioMérieux, Marcy-l’Etoile, France). To note, 25 colR isolates were from the previously described collection by Novović et al. (2017), and were included in this study for comparative purposes.
Mass Spectrometry Analysis of Lipid A
Preparations of lipid A were obtained as previously described (Kocsis et al., 2017). An aliquot of 0.7 μL of each preparation was spotted on a matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS) sample plate, mixed with an isovolume of norharmane matrix (Sigma-Aldrich, St. Louis, MO, United States) and then air-dried. Samples were analyzed with a Vitek MS instrument (bioMérieux, Marcy-l’Étoile, France) in the negative-ion mode.
DNA Extraction and Whole Genome Sequencing
Genomic DNA was extracted with the DNeasy UltraClean kit (Qiagen, Hilden, Germany), quantified by using the Qubit fluorometer (Thermo Fisher Scientific, United States) and quality checked by using the 260/280 ratio absorbance parameter as determined by the DS-11 FX + instrument (DeNovix, Wilmington, DE, United States). Sequencing was performed using a NextSeq platform (Illumina, Inc., San Diego, CA, United States) and a 2 bp × 150 bp paired-end approach. Raw data from paired-end sequencing were quality checked with the FastQC tool (v.0.11.6) and assembled with SPAdes (v.3.10.1) (Bankevich et al., 2012). One representative strain (KB-2017-139) was also sequenced with the MinION sequencer (ONT, Oxford, United Kingdom) using an R9.5.1 flow cell and the protocol 1D Genomic DNA by Ligation (SQK-LSK109). Illumina and Nanopore raw data from KB-2017-139 were assembled with a hybrid approach using Unicycler (Wick et al., 2017). Whole genome sequencing data of the 45 clinical isolates have been deposited under BioProject PRJNA4492931. The complete sequence of the plasmid pSRB_OXA-48 obtained by Illumina and Nanopore sequencing was deposited on GenBank under accession number MN218814.
Bioinformatics Analysis
MLST was performed in silico by using the tool mlst2 and the Pasteur database3. BLAST + (2.7.1) was used to detect mutations in genes potentially involved in colistin resistance (mgrB, pmrA/B, phoP/Q, crrA/B), and only mutations leading to amino acid variations were considered. For the characterization of colistin resistance mechanisms, strains of CG258, ST101 and ST336 were compared to colistin susceptible reference strains of the same CG, i.e., NJST258_2 (accession no. NZ_CP006918.1), BA33875 (NEWA00000000) and MGH-78578 (NC_009648.1), respectively. Phylogenetic relatedness was investigated with the parsnp tool (v1.2) (Treangen et al., 2014) by using default parameters and the strain NTUH-K2044 (accession no. NC_012731.1) as reference. The phylogenetic tree obtained was visualized with the online tool iTol (Letunic and Bork, 2016). The ABRicate tool4 was used to detect acquired AMR genes using the ResFinder database (Zankari et al., 2012), while plasmid replicons were predicted by PlasmidFinder (Carattoli et al., 2014). Kaptive was used for the capsular type detection (Wyres et al., 2016). Comparative analysis of plasmids was performed with BLAST Ring Image Generator (Alikhan et al., 2011) and Easyfig (Sullivan et al., 2011).
For the comparative genomic analysis of ST101 isolates, on 31 October, 2018 all the K. pneumoniae genomes available on NCBI (N = 5,820) were downloaded with the ncbi-genome-download tool5. MLST was performed and all ST101 (N = 195) (Supplementary Table S2) together with ST101 strains from this study were used for phylogenetic investigations by using parsnp and the closed ST101 chromosome from Kp_Goe_121641 (accession no. NZ_CP018735.1) as reference.
Results
Klebsiella pneumoniae Isolates and Antimicrobial Susceptibilities
In the period between November 2013 and May 2017, a total of 2,298 clinical isolates of K. pneumoniae were isolated from patients admitted to seven medical settings located in five Serbian cities. Among those, 426 isolates (18.5%) were non-susceptible to at least one carbapenem by disk diffusion, and were tested for colistin resistance. A total of 45 strains (10.6%) out of this subset showed a colistin resistant phenotype. At the time of the collection, colistin susceptibility testing was routinely performed with the Vitek2 instrument or Etest, although these methods had several limitations (Tan and Ng, 2007). Thus, the number of colR isolates may be underestimated.
All the strains were confirmed as colistin resistant by the broth microdilution method (considering the EUCAST susceptibility breakpoint of 2 mg/L) with MICs that ranged between 8 and 32 mg/L (Supplementary Table S1). Etest results for carbapenemes showed that all the strains were resistant to ertapenem, while meropenem and imipenem had susceptibility rates of 93.3 and 91.1%, respectively. Vitek2 results showed that none of the fluoroquinolones, penicillins combined with β-lactamase inhibitors and cephalosporins (including cefoxitin and the 4th generation cephalosporin cefepime) were effective against the 45 colR isolates. Conversely, amikacin (86% susceptibility) and trimethoprim/sulfamethoxazole (78% susceptibility) were the most active agents together with imipenem and meropenem (Supplementary Table S1).
Genomic Epidemiology
Genome sequence data were used to investigate the population structure of the colR K. pneumoniae strains circulating in Serbia. Five different STs were detected among the investigated collection (ST101, ST437, ST258, ST336, and ST340), with the majority of strains belonging to ST101 (N = 38) or CG258 (ST258, N = 1; ST340, N = 1 and ST437, N = 4) (Figure 1). The remaining strain belonged to CG17 and was typed as ST336. Isolates of ST101 were closely related to each other [single nucleotide polymorphism (SNP) variation: 5–893, mean 107, median 61], with only two of them (i.e., KV-2017-142 and KV-2017-143) having more than 200 SNPs when compared to other ST101 isolates and to each other. The ST101 isolates were detected in all the cities involved in this study, except Niš, thus demonstrating the endemicity at the national level of this clone. Moreover, there was not a clear clustering of isolates obtained from different hospitals, suggesting inter-hospital cross infections.
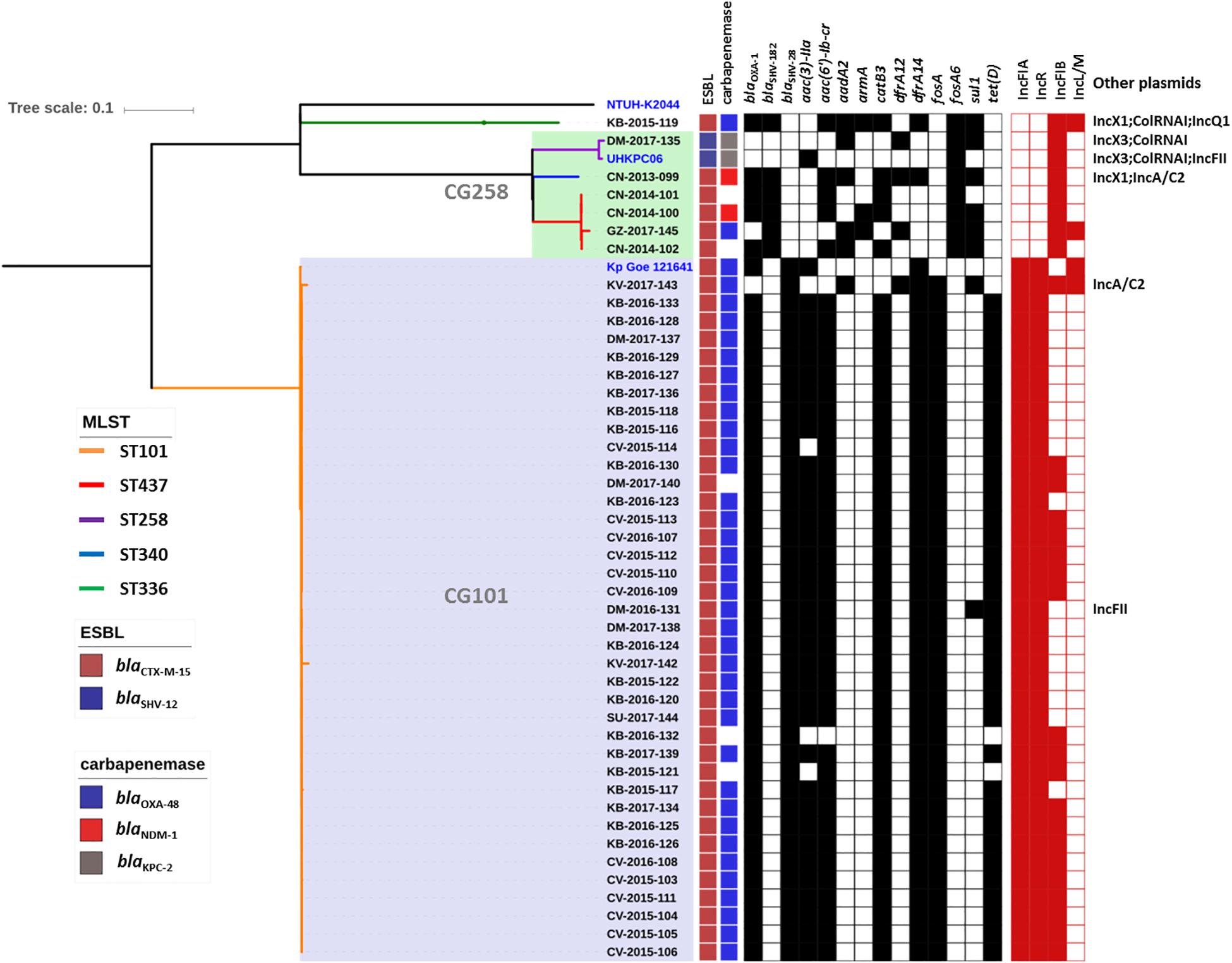
Figure 1. Phylogenetic tree of the colR Klebsiella pneumoniae isolates from Serbia. For each isolate, the medical setting (CN, Clinical center of Niš, Niš; CV, Clinical center of Vojvodina, Novi Sad; KB, Konzilijum, Belgrade; DM, University hospital center “Dr. Dragiša Mišovic-Dedinje,” Belgrade; KV, The General hospital “Studenica,” Kraljevo; GZ, The Institute of Public health of Belgrade, Belgrade; SU, General Hospital Subotica, Subotica), the year of isolation and the sample number are reported. Colored nodes indicate MLST, while the presence/absence of ESBLs, carbapenemases, resistance genes (black) and plasmid replicons is indicated by filled boxes.
The genomes of the ST101 Serbian isolates were compared with 195 ST101 genomes available in the INSDC databases, and their phylogenetic relation is showed in Figure 2. Strains from our study (red lines) cluster together in the tree in a well-defined branch containing other strains from Serbia, Slovenia, Turkey and Greece. Overall, the number of SNPs among all analyzed ST101 isolates ranged between 1 and 1,547 (mean 195, median 135), and two major lineages within this group can be observed. The majority of SNPs separating these two lineages fell in the gene cluster, and this was consistent with the previous observations that strains of ST101 are characterized by two different K-loci, KL17 and KL106, associated with wzi alleles 137 and 29, respectively (Roe et al., 2019). While KL17 is prevalent among ST101 strains, KL106 is less frequent but, interestingly, it is the second most abundant capsular variant of CG258 (Wyres et al., 2015), reinforcing the hypothesis that capsular exchange in K. pneumoniae is a common event (Chen et al., 2014; Bowers et al., 2015).
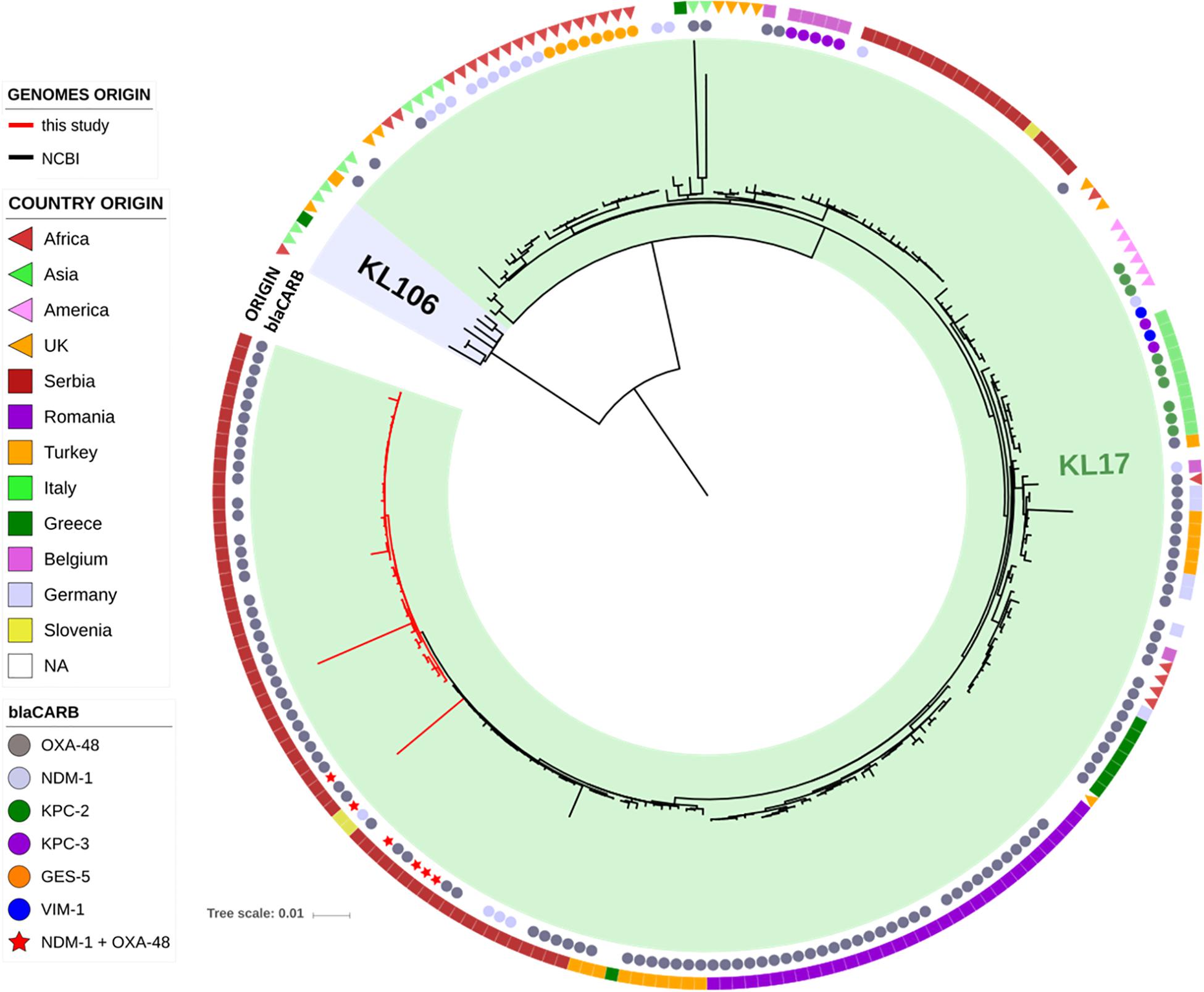
Figure 2. Phylogenetic tree of the ST101 K. pneumoniae isolates from this study (red lines) in comparison to ST101 isolates retrieved from NCBI (black lines). The two types of capsular polysaccharides (KL17 and KL106) are indicated by colored ranges. Two datasets are also present, indicating the type of carbapenemase (inner circle) and the country of origin (outer circle).
All non-ST101 isolates (excluding KB-2015-119) were part of a single monophyletic subclade within the CG258 (Bowers et al., 2015) and produced different carbapenemases or were carbapenemase negative (Figure 1), while the remaining isolate of ST336 was a OXA-48-producer and harbored the KL25 capsular type.
Colistin Resistance Mechanisms
No mcr genes were observed in the genomes of the colR isolates. Conversely, all of them showed alterations in the PhoP/PhoQ regulator mgrB gene. These alterations were mainly SNPs, with the majority of ST101 isolates from this study characterized by the mutation MgrBC28S (N = 37; 97.4%). Although different substitutions of the cysteine amino acid at position 28 have already been described (e.g., MgrBC28F and MgrBC28Y), and their role in colistin resistance has been experimentally demonstrated (Cannatelli et al., 2014b; Olaitan et al., 2014; Cheng et al., 2015; Wright et al., 2015), the MgrBC28S is first described here. This cysteine residue has been previously shown to be involved in a key disulfide bond relevant to MgrB function (Lippa and Goulian, 2012), thus its substitution by Serine or by any other amino acid is expected to interfere with the ability to repress PhoQ, leading to the overexpression of the pmrHFIJKLM operon and to a colistin resistance phenotype. The isolate CN-2013-099, belonging to ST340, displayed the previously studied MgrBC28Y substitution (Cheng et al., 2015). Different mutations leading to premature stop codons were MgrBK2* in the ST101 isolate KV-2017-143, firstly described here, MgrBK3* in the ST437 isolate GZ-2017-145 (Nordmann et al., 2016) and MgrBQ30* in the ST336 strain KB-2015-119 (Nordmann et al., 2016). The ST258 isolate was characterized by an insertion sequence of the family IS5 which interrupted the mgrB gene at nucleotide 75. Disruption of the mgrB gene by insertion sequences has been shown as a common mechanism of colistin resistance in KPC harboring strains (Cannatelli et al., 2014b). Three ST437 strains were characterized by an adenine deletion within the polyadenine region present from nucleotide 4 to 9 in mgrB, resulting in a frameshift mutation. Collectively, the results of these analyses demonstrated that all colistin resistant strains investigated in this study were characterized by genetic alterations in the mgrB gene.
Other genetic alterations potentially involved in colistin resistance were: PmrAE57G (KB-2015-119, ST336), PmrBT157P (CCV-2015-105, ST101) and PhoQV446G (CCDM-2017-135, ST258). Among these, only PmrBT157P was previously reported, and its role in reducing colistin susceptibility was demonstrated (Jayol et al., 2014). Accordingly, the ST101 isolate CV-2015-105 having PmrBT157P together with MgrBC28S, showed a colistin MIC 1- to 2-fold higher than isogenic strains carrying only MgrBC28S.
Mass spectrometry of lipid A was performed on a subset of isolates representative of the different alterations potentially involved in colistin resistance. Compared to the colistin susceptible reference ATCC11296 strain, colR isolates showed an additional peak at 1,971 m/z resulting from the addition of a 4-amino-4-deoxy-L-arabinose moiety (131 m/z) to lipid A (peak at 1,840 m/z), as previously reported (Leung et al., 2017) (results not shown). This supports the role of the observed mutations in the overexpression of the pmrHFIJKLM operon and consequent lipid A modification, leading to reduced colistin interactions. Moreover, no addition of pEtN moieties to lipid A were observed, consistently with the absence of mcr-like genes (Liu et al., 2017).
To note, our findings concerning MgrB alterations differ from those previously reported by Novović et al. (2017), as they did not detect significant MgrB alterations for most of the isolates. This underline the importance of using well-characterized colistin susceptible reference isolates, as the one used in the mentioned study was not characterized with reference methods for colistin susceptibility testing (Mirovic et al., 2012).
Other Antibiotic Resistance Mechanisms
All strains were positive for an ESBL-encoding gene, with blaCTX–M–15 harbored by all strains except the only ST258, which carried a blaSHV–12 gene. Analysis of the ompK35 gene, encoding a major outer membrane protein, showed that all non-ST258 strains had deletions leading to frameshift and premature stop codons, while the ompK36 gene was intact in all the genomes. Outer membrane impermeability most likely explains resistance to cefoxitin (a cephamycin) and to ertapenem for those isolates negative for a carbapenemase encoding gene (Ardanuy et al., 1998). Two ST437 and the ST336 isolate harbored the 16S rRNA methylase gene armA, which confers high level resistance to aminoglycosides. Several other AMR genes were observed for the following antimicrobial classes: aminoglycosides (presence of aac-, aad-, aph-, and ant-type modifying enzymes), fluoroquinolones (oqxAB, qnrB1, aac(6′)-Ib-cr, parCS80I, gyrAS83Y–S83I–D87G–D87N), phenicol (floR, catA1 and catB4 genes), sulfonamide (sul1 and sul2 genes), tetracycline (tetA and tetD genes) and trimethoprim (dfrA).
Novel IncR/IncFIA OXA-48 Plasmid Within ST101 Isolates
The production of OXA-48 was at the basis of carbapenem resistance in the K. pneumoniae of ST101 analyzed in this study. For this reason, we deeply investigated the genetic context of this gene. Spreading of the blaOXA–48-encoding gene among Enterobacterales is mainly related to the dissemination of a single ∼62-kb IncL/M-like conjugative plasmid (Poirel et al., 2012). However, PlasmidFinder analysis did not detect any IncL/M replicon among ST101 isolates from Serbia. Therefore, MinION sequencing was performed on one representative strain (KB-2017-139) with the aim to fully characterize the genomic background of the blaOXA–48 gene.
The blaOXA–48 gene was located on a plasmid of 83,654 bp, named pSRB_OXA-48, carrying both the IncR and the IncFIA type replicons, the blaCTX–M–15 and several other AMR genes (tet(D), aac(6′)-Ib-cr, blaOXA–1, catB3-like, aac(3′)-IIa and dfrA14). A BLAST analysis showed that pSRB_OXA-48 is a hybrid plasmid composed by (i) a fragment having 99.7% identity with the IncFIA-IncR pKp_Goe_641-1 plasmid (CP018737.1) and carrying the blaCTX–M–15 gene and several other AMR genes [aac(3)-IIa, catB3, blaOXA–1, aac(6′)-Ib-cr, aac(6′)-Ib, ant(3′′)-Ia, blaOXA–9, blaTEM–1A, dfrA14], and (ii) a fragment identical to the IncL/M plasmid pKp_Goe_641-2 (CP018736.1) carrying the blaOXA–48 gene (Figure 3). Both these plasmids have been described in K. pneumoniae strain Kp_Goe_121641 (accession no. NZ_CP018735.1), isolated from a refugee from North Africa hospitalized in Germany, in 2013. The latter strain belongs to ST101 and has a median of 142 SNPs (minimum 134, maximum 601) compared to the Serbian ST101 isolates from this study. Collectively these results suggest that pSRB_OXA-48 likely originated by recombination events between two plasmids within an ST101 strain related to Kp_Goe_121641. In order to elucidate the recombination mechanisms at the origin of pSRB_OXA-48, we compared this plasmid to pKp_Goe_641-1 and to pRA35 (LN864821.1), an IncL/M plasmid similar to pKp_Goe_641-2 but with an intact structure of the transposon Tn6237 carrying blaOXA–48 (Beyrouthy et al., 2014) (Figure 3). A detailed analysis showed that pSRB_OXA-48 contained a copy of Tn6237 which was disrupted by a IS26 composite transposon of 73.7 kbp sharing similarity with pKp_Goe_641-1. This hypothesis was corroborated by the presence of 8-bp target site duplication sequences (5′-GCGAATAA-3′) flanking the composite transposons regions (Figure 4). The results of reads-mapping performed against pSRB_OXA-48 using Illumina short-reads from the other ST101/OXA-48 strains was consistent with the presence of a pSRB_OXA-48-related plasmid in all the ST101/OXA-48 isolates. Non-ST101 OXA-48 strains (ST336 KB-2015-119 and ST437 GZ-2017-145) had the IncL/M replicon, while lacking the IncFIA and IncR replicons, suggesting that the blaOXA–48 gene was located in a classic IncL/M plasmid and not in a pSRB_OXA-48-like plasmid (Figure 1).
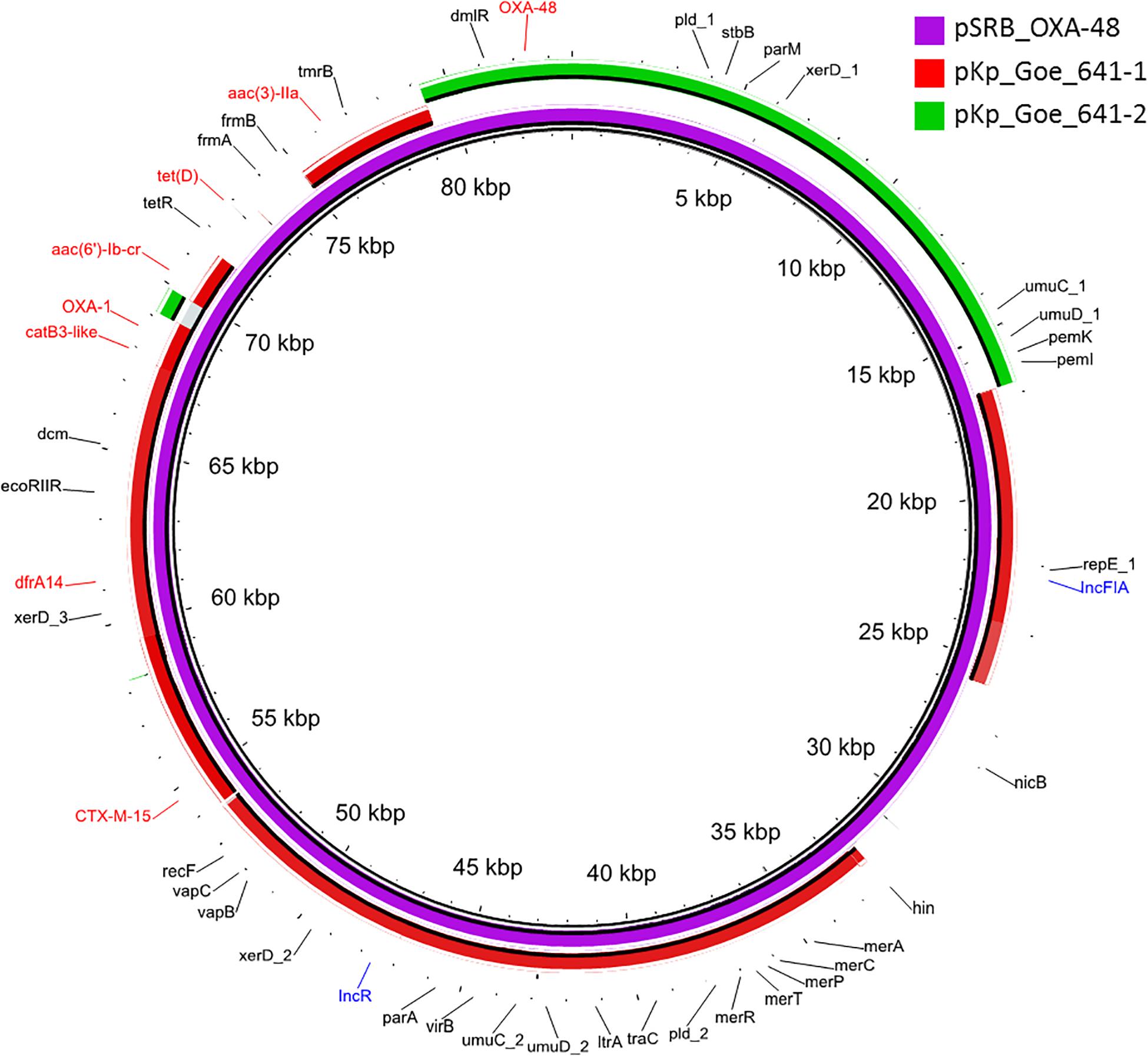
Figure 3. BLAST ring image generator output of the OXA-48 plasmid pSRB_OXA-48 from the ST101 isolate KB-2017-139 (violet) against the two major plasmids from the ST101 isolate Kp_Goe_1216141 (pKp_Goe_641-1, in red and pKp_Goe_641-2 in green). Only identities > 95% are indicated. Antimicrobial resistance genes are indicated in red, plasmid replicons in blue and all other genes in black.
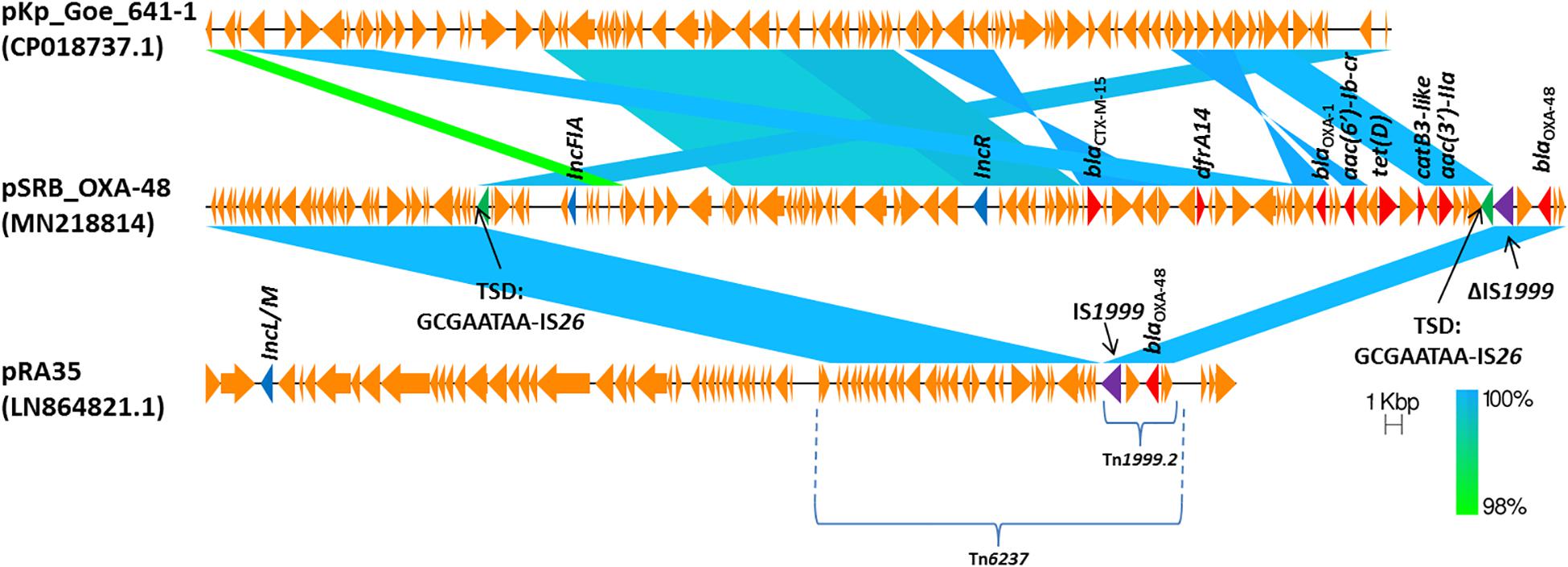
Figure 4. Comparison of plasmids pSRB_OXA-48, pKpGoe_641-1 and pRA35. Antimicrobial resistance genes, plasmid replicons and mobile elements are also indicated. TSD: target site duplication.
Discussion
This study exploited WGS to characterize a collection of colR CRKP isolates obtained from seven medical settings and five Serbian cities over a nearly 4-year period. Results showed that all the isolates presented alterations in the PhoP/PhoQ regulator MgrB, confirming its major role in colR in K. pneumoniae. Lipid A alterations associated with colR were also studied with MALDI-TOF MS. The analysis revealed the addition of a 4-amino-4-deoxy-L-arabinose moiety to lipid A, but no addition of pEtN moieties, for all isolates tested. These results support the role of the MgrB mutations in colistin resistance, and also confirm the absence of mcr-like genes.
The predominant ST observed was ST101, an emerging high-risk clone detected worldwide and associated with different carbapenemases and high mortality (Navon-Venezia et al., 2017; Can et al., 2018). In a recent European survey of CRKP isolates, including 244 hospitals in 32 countries, four major clonal lineages accounted for roughly 70% of the carbapenemase-producing isolates, including ST 11, 15, 101, 258/512 and their derivatives (David et al., 2019). The first ST101 strain from Serbia was isolated in 2013, and coproduced the OXA-48 and the NDM-1 carbapenemases (Seiffert et al., 2014). Most of the colR ST101 from this study were carbapenemase-producers, and OXA-48 was the only carbapenemase expressed. ST101/OXA-48 has been frequently reported, and in an 11-year epidemiology study of OXA-48 producers among European and north- African countries, a quarter of the OXA-48 K. pneumoniae isolates belonged to ST101 (Potron et al., 2013). Outbreaks of ST101/OXA-48 were also described, with reports from Spain (Pitart et al., 2011; Cubero et al., 2015), Algeria (Loucif et al., 2016), Czech Republic (Skálová et al., 2016) and Greece (Avgoulea et al., 2018). The challenging phenotypic detection of OXA-48 carbapenemases and the rapid horizontal transfer of OXA-48-encoding plasmids favor hospital outbreaks linked to patient transfer (Skálová et al., 2016) and draw attention to the need for continuous and meticulous surveillance, as well as timely investigation.
The blaOXA–48 gene spread is mainly related to the dissemination of a single ∼62-kb IncL/M-like conjugative plasmid that does not carry additional resistance determinants (Poirel et al., 2012). Conversely, ST101/OXA-48 isolates from this study carried a novel hybrid plasmid (pSRB_OXA-48) with replicons IncR and IncFIA and encoding OXA-48, the CTX-M-15 ESBL and several other AMR genes. Such plasmids confer an MDR phenotype which limits the use of most β-lactams, including carbapenems. In fact, even if most isolates (91%) were susceptible to imipenem, carbapenems have been proven to be not effective in an in vivo murine model (Wiskirchen et al., 2014). Moreover, there have been a number of case reports and series describing treatment failures with carbapenem-containing regimens in the treatment of OXA-48-producing bacterial infections (Stewart et al., 2018). Ceftazidime-avibactam may represent an effective alternative against such isolates, as previously reported (Kazmierczak et al., 2018).
Similarities among the Serbian ST101 strains, supported by the limited number of SNPs observed and the presence of the same alteration in the mgrB gene, suggest a clonal expansion of this clone among Serbian medical settings. This observation underscores the need to strengthen contact precautions for patients diagnosed with or suspected of having CRKP infections to limit the diffusion of colR CRKP of ST101.
Of note, colR ST101 strains have recently been associated with high mortality rates. Indeed, a prospective cohort study showed that among colR isolates, ST101 was found to be a significant independent predictor of patient mortality, with a 30 day patient mortality of 72% (Can et al., 2018).
Conclusion
This work corresponds to the first genomic investigation of colistin resistance in K. pneumoniae isolates from Serbia. The major role of MgrB mutations in colistin resistance in K. pneumoniae, observed in strains of CG258, is here confirmed for those of ST101. We also report the full sequence of a novel plasmid, pSRB_OXA-48, conferring MDR phenotype and encoding for the ESBL CTX-M-15 and the carbapenemase OXA-48.
Data Availability Statement
The datasets generated for this study can be found in the www.ncbi.nlm.nih.gov/bioproject/PRJNA449293.
Author Contributions
AB, GR, and HG conceived and designed the study. MP performed the Phenotypic and Genomics experiments. MP and MD’A performed the bioinformatics analysis. All authors analyzed the data and contributed to the manuscript.
Funding
The work performed by MP and AP was funded by the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreement No. 675412 (New Diagnostics for Infectious Diseases [ND4ID]).
Conflict of Interest
MP, AP, CM, and AB are employees of bioMérieux, a company that develops and sells diagnostic tests in the field of infectious diseases.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00294/full#supplementary-material
Footnotes
- ^ www.ncbi.nlm.nih.gov/bioproject/PRJNA449293
- ^ https://github.com/tseemann/mlst
- ^ https://bigsdb.pasteur.fr/
- ^ https://github.com/tseemann/abricate
- ^ https://github.com/kblin/ncbi-genome-download
References
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Ardanuy, C., Liñares, J., Domínguez, M. A., Hernández-Allés, S., Benedí, V. J., and Martínez-Martínez, L. (1998). Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob. Agents Chemother. 42, 1636–1640. doi: 10.1128/aac.42.7.1636
Avgoulea, K., Di Pilato, V., Zarkotou, O., Sennati, S., Politi, L., Cannatelli, A., et al. (2018). Characterization of extensively- or pandrug-resistant ST147 and ST101 OXA-48-producing Klebsiella pneumoniae isolates causing bloodstream infections in ICU patients. Antimicrob. Agents Chemother. 62:e2457-e2417. doi: 10.1128/AAC.02457-17
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bassetti, M., Righi, E., Carnelutti, A., Graziano, E., and Russo, A. (2018). Multidrug-resistant Klebsiella pneumoniae?: challenges for treatment, prevention and infection control. Expert Rev. Anti Infect. Ther. 16, 749–761. doi: 10.1080/14787210.2018.1522249
Beyrouthy, R., Robin, F., Delmas, J., Gibold, L., Dalmasso, G., Dabboussi, F., et al. (2014). IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of blaOXA-48 into the Escherichia coli chromosome. Antimicrob. Agents Chemother. 58, 3785–3790. doi: 10.1128/AAC.02669-14
Bowers, J. R., Kitchel, B., Driebe, E. M., MacCannell, D. R., Roe, C., Lemmer, D., et al. (2015). Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 10:e0133727. doi: 10.1371/journal.pone.0133727
Can, F., Menekse, S., Ispir, P., Atac, N., Albayrak, O., Demir, T., et al. (2018). Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 73, 1235–1240. doi: 10.1093/jac/dkx532
Cannatelli, A., D’Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Cannatelli, A., Di Pilato, V., Giani, T., Arena, F., Ambretti, S., Gaibani, P., et al. (2014a). In vivo evolution to Colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob. Agents Chemother. 58, 4399–4403. doi: 10.1128/AAC.02555-14
Cannatelli, A., Giani, T., D’Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014b). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/AAC03110-14
Carattoli, A., Zankari, E., Garciá-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Mathema, B., Pitout, J. D. D., DeLeo, F. R., and Kreiswirth, B. N. (2014). Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. mBio 5:e01355-14. doi: 10.1128/mBio.01355-1314
Cheng, H.-Y., Chen, Y.-F., and Peng, H.-L. (2010). Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 17:60. doi: 10.1186/1423-0127-17-60
Cheng, Y. H., Lin, T. L., Pan, Y. J., Wang, Y. P., Lin, Y. T., and Wang, J. T. (2015). Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob. Agents Chemother. 59, 2909–2913. doi: 10.1128/AAC.04763-14
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing, 29th Edn, CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standars Institute.
Cubero, M., Cuervo, G., Dominguez, M. Á., Tubau, F., Martí, S., Sevillano, E., et al. (2015). Carbapenem-resistant and carbapenem-susceptible isogenic isolates of Klebsiella pneumoniae ST101 causing infection in a tertiary hospital. BMC Microbiol. 15:177. doi: 10.1186/s12866-015-0510-9
David, S., Reuter, S., Harris, S. R., Glasner, C., Feltwell, T., Argimon, S., et al. (2019). Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919–1929. doi: 10.1038/s41564-019-0492-8
EUCAST (2019). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. Available online at: http://www.eucast.org (accessed June 12, 2019).
Grundmann, H., Glasner, C., Albiger, B., Aanensen, D. M., Tomlinson, C. T., Andrasević, A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163. doi: 10.1016/S1473-3099(16)30257-2
Jayol, A., Poirel, L., Brink, A., Villegas, M. V., Yilmaz, M., and Nordmann, P. (2014). Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58, 4762–4766. doi: 10.1128/AAC.00084-14
Karaiskos, I., Souli, M., Galani, I., and Giamarellou, H. (2017). Colistin: still a lifesaver for the 21st century? Expert Opin. Drug Metab. Toxicol. 13, 59–71. doi: 10.1080/17425255.2017.1230200
Kazmierczak, K. M., Bradford, P. A., Stone, G. G., de Jonge, B. L. M., and Sahm, D. F. (2018). In vitro activity of ceftazidime-avibactam and aztreonam-avibactam against OXA-48-carrying Enterobacteriaceae isolated as part of the international network for optimal resistance monitoring (INFORM) global surveillance program from 2012 to 2015. Antimicrob. Agents Chemother. 62:e592-e518. doi: 10.1128/AAC.00592-18
Kocsis, B., Kilár, A., Péter, S., Dörnyei, Á., Sándor, V., and Kilár, F. (2017). Mass spectrometry for profiling LOS and lipid a structures from whole-cell lysates: directly from a few bacterial colonies or from liquid broth cultures. Methods Mol. Biol. 1600, 187–198. doi: 10.1007/978-1-4939-6958-6_17
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Leung, L. M., Fondrie, W. E., Doi, Y., Johnson, J. K., Strickland, D. K., Ernst, R. K., et al. (2017). Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci. Rep. 7:6403. doi: 10.1038/s41598-017-04793-4
Lippa, A. M., and Goulian, M. (2012). Perturbation of the oxidizing environment of the periplasm stimulates the PhoQ/PhoP system in Escherichia coli. J. Bacteriol. 194, 1457–1463. doi: 10.1128/JB.06055-11
Liu, Y.-Y., Chandler, C. E., Leung, L. M., McElheny, C. L., Mettus, R. T., Shanks, R. M. Q., et al. (2017). Structural modification of lipopolysaccharide conferred by mcr-1 in gram-negative ESKAPE pathogens. Antimicrob. Agents Chemother. 61:e580-e517. doi: 10.1128/AAC.00580-17
Loucif, L., Kassah Laouar, A., Saidi, M., Messala, A., Chelaghma, W., and Rolain, J.-M. (2016). Outbreak of OXA-48-producing Klebsiella pneumoniae involving an ST 101 clone in Batna University Hospital, Algeria. Antimicrob. Agents Chemother. 60, 7494–7497. doi: 10.1128/AAC.00525-16
Mirovic, V., Tomanovic, B., Lepsanovic, Z., Jovcic, B., and Kojic, M. (2012). Isolation of Klebsiella pneumoniae producing NDM-1 metallo-β-lactamase from the urine of an outpatient baby boy receiving antibiotic prophylaxis. Antimicrob. Agents Chemother. 56, 6062–6063. doi: 10.1128/AAC.00838-12
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Nordmann, P., Jayol, A., and Poirel, L. (2016). Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg. Infect. Dis. 22, 1038–1043. doi: 10.3201/eid2206.151840
Novović, K., Trudić, A., Brkić, S., Vasiljević, Z., Kojić, M., Medić, D., et al. (2017). Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob. Agents Chemother. 61:e2550-e2516. doi: 10.1128/AAC.02550-16
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pitart, C., Solé, M., Roca, I., Fàbrega, A., Vila, J., and Marco, F. (2011). First Outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 β-lactamase in Klebsiella pneumoniae in Spain. Antimicrob. Agents Chemother. 55, 4398–4401. doi: 10.1128/AAC.00329-311
Poirel, L., Bonnin, R. A., and Nordmann, P. (2012). Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56, 559–562. doi: 10.1128/AAC.05289-5211
Potron, A., Poirel, L., Rondinaud, E., and Nordmann, P. (2013). Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 18:20549. doi: 10.2807/1560-7917.es2013.18.31.20549
Roe, C. C., Vazquez, A. J., Esposito, E. P., Zarrilli, R., and Sahl, J. W. (2019). Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front. Microbiol. 10:542. doi: 10.3389/fmicb.2019.00542
Seiffert, S. N., Marschall, J., Perreten, V., Carattoli, A., Furrer, H., and Endimiani, A. (2014). Emergence of Klebsiella pneumoniae co-producing NDM-1, OXA-48, CTX-M-15, CMY-16, qnrA and ArmA in Switzerland. Int. J. Antimicrob. Agents 44, 260–262. doi: 10.1016/j.ijantimicag.2014.05.008
Skálová, A., hudějová, K., Rotová, V., Medvecky, M., Študentová, V., Chudáčková, E., et al. (2016). Molecular characterization of OXA-48-like-producing Enterobacteriaceae in the Czech Republic: evidence for horizontal transfer of pOXA-48-like plasmids. Antimicrob. Agents Chemother. 61:e01889-16. doi: 10.1128/AAC.01889-16
Stewart, A., Harris, P., Henderson, A., and Paterson, D. (2018). Treatment of infections by OXA-48-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 62:e01195-18. doi: 10.1128/AAC.01195-18
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Zhang, H., Liu, Y.-H., and Feng, Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Tan, T. Y., and Ng, S. Y. (2007). Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clin. Microbiol. Infect. 13, 541–544. doi: 10.1111/j.1469-0691.2007.01708.x
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Trudic, A., Jelesic, Z., Mihajlovic-Ukropina, M., Medic, D., Zivlak, B., Gusman, V., et al. (2017). Carbapenemase production in hospital isolates of multidrug-resistant Klebsiella pneumoniae and Escherichia coli in Serbia. Vojnosanit. Pregl. 74, 715–721. doi: 10.2298/VSP150917260T
WHO Regional Office for Europe (2017). Central Asian and Eastern European Surveillance of Antimicrobial Resistance. Copenhagen: WHO Regional Office for Europe.
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wiskirchen, D. E., Nordmann, P., Crandon, J. L., and Nicolau, D. P. (2014). Efficacy of humanized carbapenem and ceftazidime regimens against Enterobacteriaceae producing OXA-48 carbapenemase in a murine infection model. Antimicrob. Agents Chemother. 58, 1678–1683. doi: 10.1128/AAC.01947-13
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-4014
Wyres, K. L., Gorrie, C., Edwards, D. J., Wertheim, H. F. L., Hsu, L. Y., Van Kinh, N., et al. (2015). Extensive capsule locus variation and large-scale genomic recombination within the Klebsiella pneumoniae clonal group 258. Genome Biol. Evol. 7, 1267–1279. doi: 10.1093/gbe/evv062
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2:e000102. doi: 10.1099/mgen.0.000102
Keywords: blaOXA–48, K. pneumoniae, colistin, mgrB, Serbia, ST101, WGS
Citation: Palmieri M, D’Andrea MM, Pelegrin AC, Mirande C, Brkic S, Cirkovic I, Goossens H, Rossolini GM and van Belkum A (2020) Genomic Epidemiology of Carbapenem- and Colistin-Resistant Klebsiella pneumoniae Isolates From Serbia: Predominance of ST101 Strains Carrying a Novel OXA-48 Plasmid. Front. Microbiol. 11:294. doi: 10.3389/fmicb.2020.00294
Received: 09 December 2019; Accepted: 10 February 2020;
Published: 21 February 2020.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Jesús Mingorance, University Hospital La Paz, SpainPaolo Gaibani, Sant’Orsola-Malpighi Polyclinic, Italy
Copyright © 2020 Palmieri, D’Andrea, Pelegrin, Mirande, Brkic, Cirkovic, Goossens, Rossolini and van Belkum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alex van Belkum, alex.vanbelkum@biomerieux.com
 Mattia Palmieri
Mattia Palmieri Marco Maria D’Andrea
Marco Maria D’Andrea Andreu Coello Pelegrin
Andreu Coello Pelegrin Caroline Mirande4
Caroline Mirande4 Snezana Brkic
Snezana Brkic Gian Maria Rossolini
Gian Maria Rossolini Alex van Belkum
Alex van Belkum