- 1Laboratory of Quality and Safety Risk Assessment for Dairy Products of Ministry of Agriculture and Rural Affairs, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Key Laboratory of Quality & Safety Control for Milk and Dairy Products of Ministry of Agriculture and Rural Affairs, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
Escherichia coli is a common bacterium in the intestines of animals, and it is also the major important cause of toxic mastitis, which is an acute or peracute disease that causes a higher incidence of death and culling of cattle. The purpose of this study was to investigate E. coli strains isolated from the raw milk of dairy cattle in Northern China, and the antibacterial susceptibility of these strains and essential virulence genes. From May to September 2015, 195 raw milk samples were collected from 195 dairy farms located in Northern China. Among the samples, 67 (34.4%) samples were positive for E. coli. About 67 E. coli strains were isolated from these 67 samples. The prevalence of Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), and enteroinvasive E. coli (EIEC) were 9, 6, 4.5, and 1.5%, respectively. Among the virulence genes detected, stx1 was the most prevalent (6/67, 9%) gene, followed by eae (3/67, 4.5%), and estB (2/67, 3%). Moreover, the strains exhibited different resistance levels to ampicillin (46.3%), amoxicillin-clavulanic acid (16.4%), trimethoprim-sulfamethoxazole (13.4%), tetracycline (13.4%), cefoxitin (11.9%), chloramphenicol (7.5%), kanamycin (7.5%), streptomycin (6.0%), tobramycin (4.5%), azithromycin (4.5%), and ciprofloxacin (1.5%). All of the E. coli isolates were susceptible to gentamicin. The prevalence of β-lactamase-encoding genes was 34.3% in 67 E. coli isolates and 45% in 40 β-lactam-resistance E. coli isolates. The overall prevalence of blaSHV, blaTEM, blaCMY, and blaCTX-M genes were 1.5, 20.9, 10.4, and 1.5%, respectively. Nine non-pathogenic E. coli isolates also carried β-lactamase resistance genes, which may transfer to other pathogenic E. coli and pose a threat to the farm’s mastitis management projects. Our results showed that most of E. coli were multidrug resistant and possessed multiple virulence genes, which may have a huge potential hazard with public health, and antibiotic resistance of E. coli was prevalent in dairy herds in Northern China, and ampicillin should be used cautiously for mastitis caused by E. coli in Northern China.
Introduction
Escherichia coli was a common inhabitant of the intestine of animals (Tark et al., 2016). During parturition and early lactation period, E. coli was found to usually infect mammary gland of cows, which may cause acute and local mastitis (Hinthong et al., 2017). Escherichia coli is the main cause of bacterial mastitis in cows. It is usually short-lived, causing the infection that lasts 2–3days. However, E. coli has been displayed to cause persistent infections in a few cases (Lippolis et al., 2017). Pathogenic E. coli can cause disease in animals and humans due to virious virulence (Ntuli et al., 2016). Based on the epidemiological, clinical, and pathogenic characteristics, E. coli is classified into different pathotypes: Shiga toxin-producing E. coli (STEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), and enteroinvasive E. coli (EIEC; Rugeles et al., 2010). Numerous outbreaks associated with E. coli in milk and other foods have been reported recently (EFSA-ECDC, 2012; EFSA, 2015; Ombarak et al., 2016). For example, STEC can generate two types of Shiga toxins (stx1 and stx2), and EPEC can produce bfp gene, which were involved in pathogenicity of gastrointestinal tract (Hernandes et al., 2009; Douellou et al., 2016). ETEC can express heat-stable est genes that can cause severe diarrhea. EAEC can produce aggR gene, which were associated with the generation of biofilm (Medeiros et al., 2013). The ipaH gene from EIEC can lead to the occurrence of fever, vomiting, and dehydration in infected children. The higher prevalence of E. coli is closely associated with hygiene in raw milk (Radostits et al., 2007). Therefore, the study on E. coli in raw milk is significant.
Escherichia coli is not only with the potential occurrence, but also with the rapid development of antibiotic resistance bacteria (Ntuli et al., 2016). Inappropriate selection and abuse of antibiotics could lead to antibiotic resistance in bacteria (Da Silva and Mendonça, 2012). Moreover, E. coli may develop acquired resistance to other antibiotics by carrying various resistance characteristics on mutation, plasmids, or transposons (Gonggrijp et al., 2016). For example, extended-spectrum β-lactamases E. coli, resistant to β-lactam antibiotics including third- and fourth-generation cephalosporins, acquires ESBL by mutation or plasmid-mediated horizontal gene transfer (Freitag et al., 2016). Acquired antibiotic resistance also has a transmission potential to humans and other animals (Ruegg et al., 2014). Raw milk can also facilitate the transmission of antibiotic resistance genes to the human gastrointestinal tract, In addition to the presence of pathogenic bacteria. A better understanding on the resistance profile of E. coli isolates will improve our understanding of appropriate treatments for pathogen-related management (Tark et al., 2016). Therefore, monitoring the antibiotic resistance of E. coli in raw milk may show the trend or specific characteristics of antibiotic resistance and help to better prevent or more effectively treat mastitis on dairy farm.
Antimicrobial resistance and virulence types in E. coli have been studied on raw milk of healthy dairy cattle and of bovine mastitis in a variety of countries, including Northern Italy, Romania, Brazil, Egypt, South Korea, and Thailand (Trevisani et al., 2014; Ombarak et al., 2016; Ribeiro et al., 2016; Tark et al., 2016; Hinthong et al., 2017; Tabaran et al., 2017). However, incidence on antibiotic resistance of E. coli from raw milk in Northern China were very limited. Continuous monitoring of the antibiotic resistance and virulence type of E. coli could be necessary to evaluate E. coli risk in raw milk. Therefore, the objective of the work was to investigate the rate of E. coli strains isolated from raw milk in Northern China, and to characterize the antimicrobial susceptibility and key virulence genes of these strains.
Materials and Methods
Collection of Samples
In total, 195 raw milk were collected from 195 dairy farms from four cities, which was the major dairy-production cities of Northern China (herd size ≥300, no clinical mastitis cow, milking frequency two or three times per day), from May to September in 2015 (average daily temperature >20°C). There were 30 raw milk samples from Jinan, 40 samples from Harbin, 50 samples from Beijing, and 75 samples from Hohhot (Figure 1). The raw milk samples were collected from the top, middle, and bottom of bulk tank, mixed well, and then transferred into sterile bottles and transported to laboratory at 4°C immediately.
Figure 1. Map of sampling locations. In total, 195 samples were collected from Hohhot, Beijing, Harbin, and Jinan.
Isolation and Identification of E. coli
Aliquots (25ml) of each sample were added to 225ml tryptic soy broth, and then incubated at 37°C for 16h with shaking for E. coli detection. The samples were placed onto Eosin Methylene Blue agar plates (Beijing Land Bridge Technology Ltd., Beijing, China). The agar plates were incubated at 37°C for 18–24h. The presumptive colonies (typical blue-black appearance with a metallic green sheen) were picked. All the colonies were sub-cultured onto nutrient agar slants at 37°C for 16h, and then used for biochemical identification. The colonies initially identified as E. coli were examined by Voges-Proskauer negative, methyl-red positive and citrate negative. All isolates were stored at −80°C until use.
All the presumptive colonies were confirmed by PCR on 16S rRNA gene detection (Supplementary Table S1). Genomic DNA was extracted with the InstaGene Matrix DNA extraction kit (Bio-Rad Laboratories), based on the manufacturer’s instruction. PCR were performed with the EmeraldAmp Max PCR Master Mix kit (Takara, Dalian, China) followed the instructions of manufacturer. The primers were synthesized by GeneCreate Biological Engineering Co., Ltd. (Wuhan, China). Briefly, 25μl reactions, which contains 12.5μl of 2×EmeraldAmp Max PCR Master Mix kit, 10pmol of each primer, 1μl of extracted DNA and ultrapure water, were prepared. The amplification conditions were as follows: 94°C for 3min; 30cycles of 94°C for 30s, 55°C for 30s, and 72°C for 1min; and 72°C for 10min for a final extension step. Without genomic DNA as negative control and E. coli ATCC 25922 as positive control were included in all the PCR assays.
Detection of Virulence Determinants
Seven virulence genes for each diarrheagenic E. coli were detected by PCR method: stx1 and stx2 for STEC, estA, estB, and eltB for ETEC, aggR for EAEC, bfp and eae for EPEC, and ipaH for EIEC. Amplified products were analyzed by agarose gel electrophoresis, and then visualized by SYBR Safe DNA Stain gel staining. All the primers were shown in Supplementary Table S1.
Antimicrobial Susceptibility Patterns
Antimicrobial patterns for recovered E. coli were determined by agar disk diffusion method (CLSI, 2012). Gentamicin (10μg), tobramycin (10μg), streptomycin (10μg), amoxicillin-clavulanic acid (20/10μg), ampicillin (10μg), ciprofloxacin (5μg), azithromycin (15μg), cefoxitin (30μg), chloramphenicol (30μg), tetracycline (30μg), kanamycin (30μg), and trimethoprim-sulfamethoxazole (1.25/23.75μg) were used as antibiotic agents (Oxoid, Basingstoke, United Kingdom). Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 6538 were used as quality controls. The experiment was repeated three times.
Antimicrobial Resistance Genes
Four β-lactamase resistance related genes (blaCMY, blaSHV, blaCTX-M, and blaTEM) and two tetracycline genes (tetA and tetB) were detected by multiplex PCR in E. coli strains (Supplementary Table S1). The amplification conditions were as follows: 95°C for 5min, 30cycles of 94°C for 30s, 63°C for 90s, and 72°C for 90s, and 72°C for 7min for a final extension step (Ribeiro et al., 2016). Escherichia coli strains ATCC 25922 was used as a positive control in each run.
Results
Prevalence of E. coli
Out of 195 samples, 67 (34.4%) raw milk samples were positive for E. coli. Among these 67 raw milk samples, 67 E. coli strains were isolated, including 11 strains (36.7%) of 30 Jinan samples, 23 strains (30.7%) of 75 Hohhot samples, 16 strains (40.0%) of 40 Harbin samples, and 17 strains (34.0%) of 50 Beijing samples.
Virulence Genes
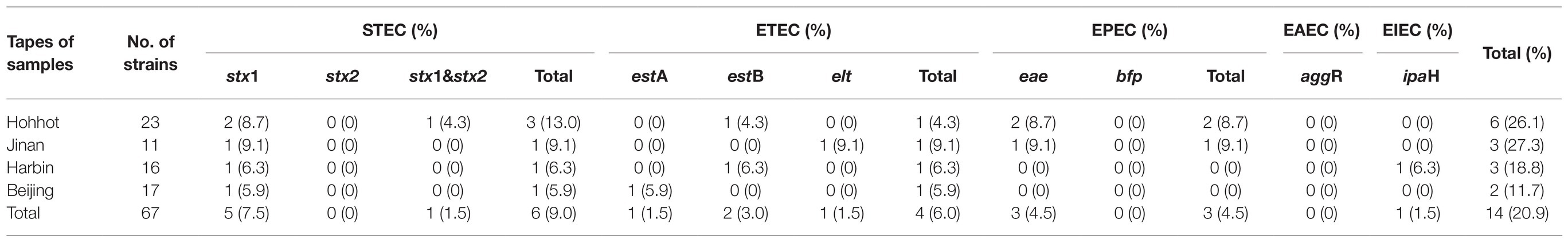
About 20.9% of the isolates (14/67) harbored more than one virulence gene, as shown in Table 1. The prevalence of EAEC, EIEC, EPEC, ETEC, and STEC was 0, 1.5, 4.5, 6, and 9%. Among the virulence genes detected, stx1 was the most prevalent gene (6/67, 9%), followed by eae (3/67, 4.5%), estB (2/67, 3%), stx2 (1/67, 1.5%), estA (1/67, 1.5%), elt (1/67, 1.5%), and ipaH (1/67, 1.5%). The aggR and bfp were not discovered in any E. coli strains. Among six STEC isolates, there were three isolates (13.0%) from Hohhot, one isolate (9.1%) from Jinan, one isolate (6.3%) from Harbin, and one isolate (5.9%) from Beijing, respectively. There were two eae-positive isolates from Hohhot (8.7%), one eae-positive isolates from Jinan (9.1%), and no eae-positive isolate from Harbin and Beijing. Moreover, the prevalence of ETEC strains were 9.1% from Jinan, 6.3% from Harbin, 5.9% from Beijing, and 4.3% from Hohhot, respectively. For ETEC-related virulence genes, the prevalence of estB, estA, and elt genes were 3.0% (2/67), 1.5% (1/67), and 1.5% (1/67), and there were one elt-positive isolate from Jinan (9.1%), two estB-positive isolates from Harbin (6.3%) and Hohhot (4.3%), and one estA-positive isolate from Beijing (5.9%). The ipaH was detected in only one E. coli strain from Harbin.
Antimicrobial Susceptibility Testing
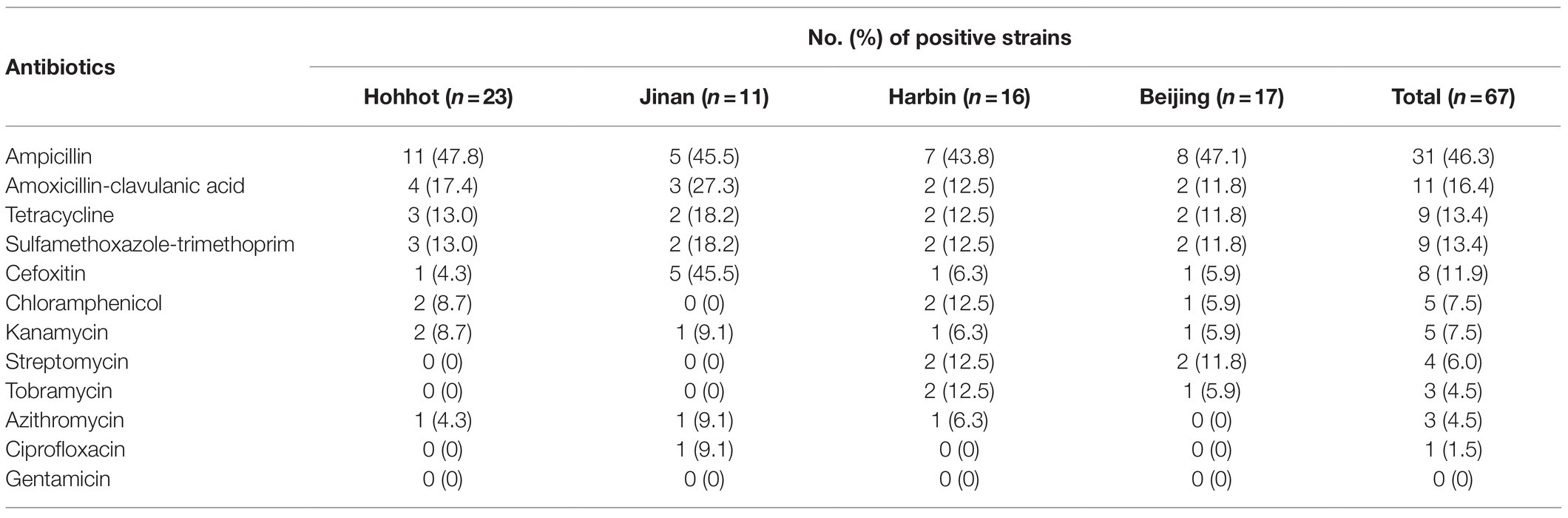
The 67 isolates were exanimated by the disk diffusion method for susceptibility to 12 antibiotics. Antibiotic resistance on E. coli was observed to ampicillin (46.3%), amoxicillin-clavulanic acid (16.4%), tetracycline (13.4%), trimethoprim-sulfamethoxazole (13.4%), cefoxitin (11.9%), chloramphenicol (7.5%), kanamycin (7.5%), streptomycin (6.0%), tobramycin (4.5%), azithromycin (4.5%), ciprofloxacin (1.5%), and gentamicin (0; Table 2). Among isolates from Hohhot, the resistant to ampicillin (47.8%) was the most frequently observed, followed by amoxicillin-clavulanic acid (17.4%), tetracycline (13.0%), and sulfamethoxazole-trimethoprim (13.0%), and all investigated strains were sensitive to tobramycin, streptomycin, ciprofloxacin, and gentamicin. Among isolates from Jinan, the resistance to ampicillin and cefoxitin (45.5%) was the most frequently observed, and all investigated strains were sensitive to tobramycin, streptomycin, and gentamicin. Among isolates from Harbin, the resistance to ampicillin (43.8%) was the most frequently observed, followed by amoxicillin-clavulanic acid, tetracycline, sulfamethoxazole-trimethoprim, chloramphenicol, streptomycin, and tobramycin (12.5%), and all investigated strains were sensitive to ciprofloxacin and gentamicin. Among isolates from Beijing, the resistance to ampicillin (47.1%) was the most frequently observed, followed by streptomycin (12.5%), amoxicillin-clavulanic acid (11.8%), tetracycline (11.8%), and sulfamethoxazole-trimethoprim (11.8%), and all the investigated E. coli isolates were sensitive to ciprofloxacin, azithromycin, and gentamicin. Moreover, 38 strains (71.6%) were resistant to at least one antibiotic, and 13 isolates (19.4%) were resistant to more than three kinds of antibiotics.
Screening of Antibiotic Resistance Genes
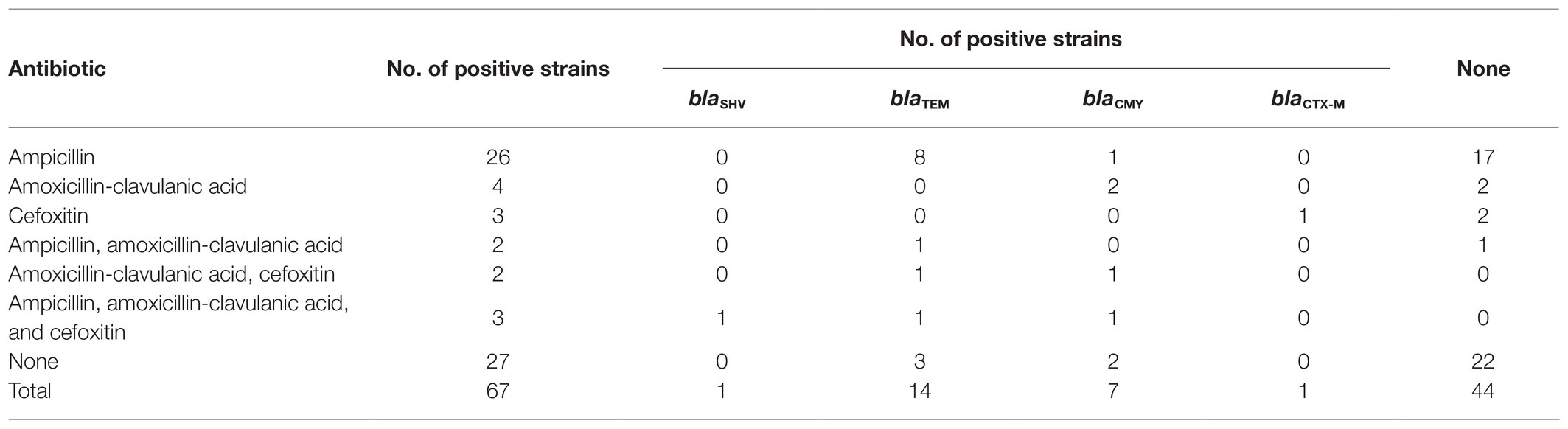
The β-lactamase-encoding genes results were presented in Table 3. The prevalence of β-lactamase-encoding genes were 34.3% in 67 E. coli isolates and 45% in 40 β-lactam resistance E. coli isolates. The overall prevalences of blaSHV, blaTEM, blaCMY, and blaCTX-M genes among E. coli isolates, which was narrow spectrum extended-spectrum β-lactamase-encoding genes, β-lactamase-encoding genes, AmpC, and β-lactamase-encoding genes, were 1.5, 1.5, 10.4, and 20.9%, respectively. In total, 71.4% of the isolates, which possessed the blaTEM gene, were resistant to ampicillin. Around 57.1% of blaCMY positive isolates were resistant to amoxicillin-clavulanic acid. Five (7.5%) isolates possessing blaTEM or blaCMY did not suggest β-lactamase antibiotic resistance.
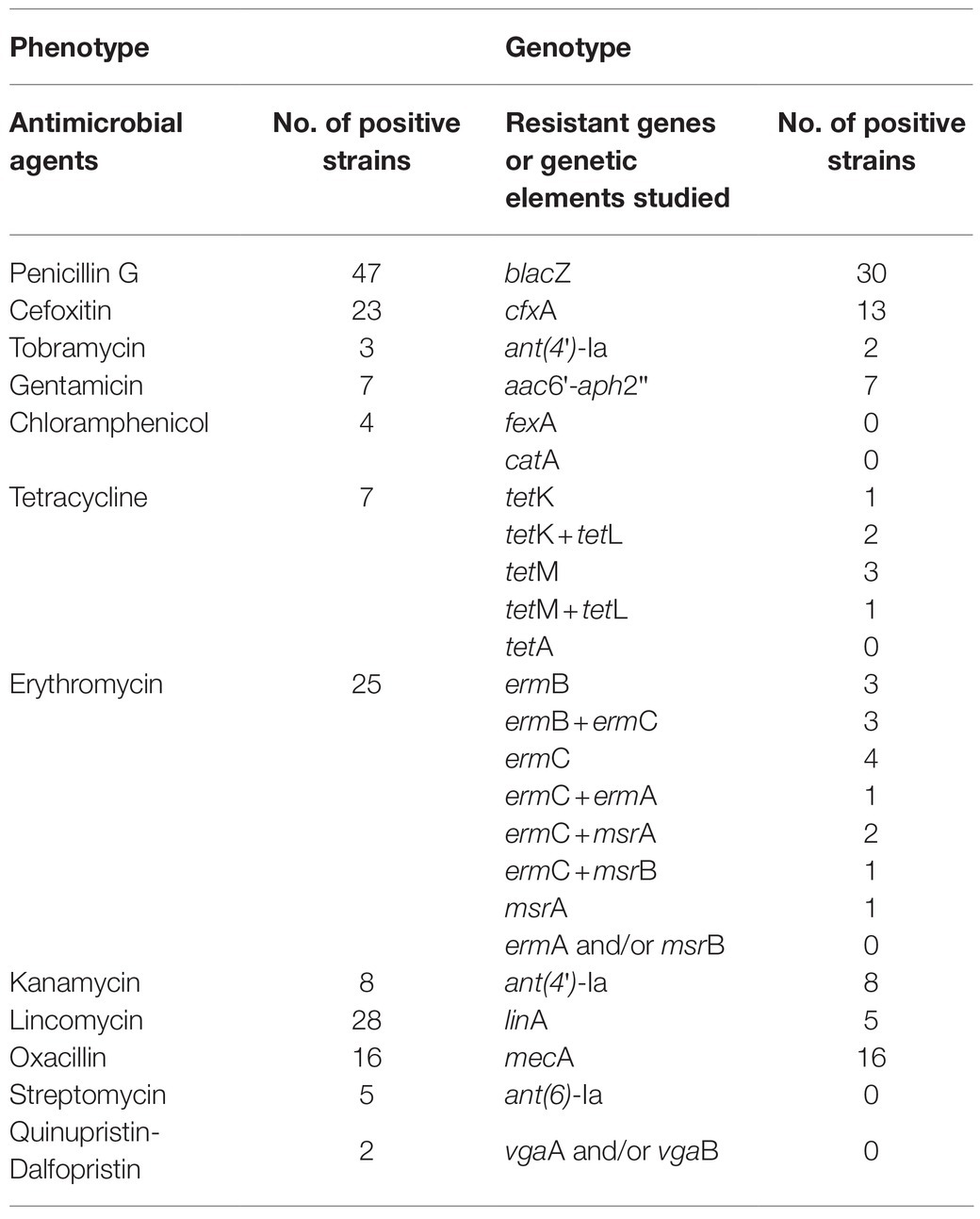
Moreover, the presence of the tet genes, which were conferring resistance to tetracycline, were confirmed in seven tetracycline-resistance strains. None of the studied strains possessed tetA (Table 4).
Discussion
In this research, 34.4% (67/195) of samples were positive for E. coli in raw milk. These results are significantly lower than that in previous studies. The incidence of E. coli in raw milk in India was 81.1% (Bhoomika et al., 2016), 75% in Bangladesh (Islam et al., 2016), 64.5% in Malaysia (Jayarao and Henning, 2001), and 45% in Northern China (Lan et al., 2017). In contrast, a much lower incidence (22.4%) of E. coli was discovered in raw milk in Sharkia Governorate (Awadallah et al., 2016). Moreover, our results are comparable with the findings of Ntuli et al. (2016), who reported 36% prevalence rate in bulk milk in South Africa, and Sharma et al. (2015), who reported 35.63% occurrence rate in raw milk in the Jaipur city of Rajasthan. Overall, the results indicated that E. coli is a common strain in raw milk collected from dairy herds of Northern China. The high prevalence of E. coli in raw milk and dairy products is a cause of concern because it is related to contamination from fecal sources and the consequent risk of enteric pathogenic microorganisms in food (Ombarak et al., 2016).
An important factor of E. coli infections is virulence factors. When E. coli carried some virulence genes, they could be potentially harmful to public consumers (Hinthong et al., 2017). In the study, 20.9% (14/67) of the tested raw milk possessing more than one virulence gene tested, may carried potentially pathogenic E. coli, as shown in Table 3. STEC, cause a life-threatening sequel, such as neurological disorder and hemolytic syndrome or HUS (Kaper et al., 2004), was found to be the most common pathogenic E. coli strain in raw milk. It has been reported that the virulence genes of STEC isolates were commonly implicated in many foodborne STEC outbreaks in the world (Beutin and Fach, 2015). In this study, the most common virulence genes in raw milk samples in Northern China were stx genes. The result was in agreement with Suojala et al. (2011), who reported the STEC (stx-positive isolates) was the most common E. coli type of raw milk with subclinical mastitis in Southern Finland, and by Lambertini et al. (2015), who found that the most frequently detected gene in raw milk of the United States northeastern was stx1. However, STEC or stx factors has been detected in the farms of United States and European at a low prevalence (Jayarao et al., 2006; Pradel et al., 2008; Van Kessel et al., 2011; Claeys et al., 2013; Ombarak et al., 2016).
Enteropathogenic E. coli is responsible for diarrhea in both developing and developed countries. As an important foodborne pathogen, EPEC has high isolation rate in retail foods in China (Zhang et al., 2016). EPEC were isolated from many animals, such as cattle, goat, sheep, chicken, gull, and pigeon (Gomez-Aldapa et al., 2016). In the study, three strains were eae genes-positive and bfp gene-negative, which could be classified as EPEC. Cortés et al. (2005) and Gomez-Aldapa et al. (2016) found that atypical EPEC strains were found in raw milk in Egypt, Saudi Arabia, and Slovakia. However, there is no report on the eae-positive E. coli strains found in mastitis cows in Iran and Thailand (Ghanbarpour and Oswald, 2010; Hinthong et al., 2017). Moreover, an increasing frequency of eae-negative isolates were postulated to have other putative adherence and virulence associated factors (Gomez-Aldapa et al., 2016). ETEC strains are usually transmitted by contaminated food. In the study, EPEC and ETEC strains were isolated from Hohhot and Jinan. EPEC/ETEC hybrid isolates were related to EPEC strain, and appeared to have acquired virulence genes by horizontal gene transfer (Hazen et al., 2017).
In the study, antimicrobial resistance was most frequently observed to ampicillin (46.3%). The susceptibility to amoxicillin can be predicted by antimicrobial resistance to ampicillin (CLSI, 2012). So the tested E. coli isolates may showed a high resistance to amoxicillin. Nam et al. (2009) reported that 32.2% E. coli strains from mastitis cow were resistant to ampicillin. However, the resistant rates in the study were much higher than those in South Korea from 2012 to 2015 (Tark et al., 2016) and in Northern Colorado (McConnel et al., 2016). Antibiotic susceptibility of E. coli was more important on choosing a suitable antibiotic for mastitis (Wang et al., 2016). The information of antibiotic use for dairy in Northern China has been investigated in our previous survey. Ampicillin was commonly used in dairy mastitis therapy (Liu et al., 2017). So, ampicillin is not a suitable treatment for mastitis caused by E. coli in Northern China.
In our previous survey, we found that five antibiotics (penicillin, ciprofloxacin, sulfamethoxazole-trimethoprim, streptomycin, and gentamicin) were commonly used in mastitis cow. In the study, most of tested strains showed an obvious antimicrobial resistance to ciprofloxacin, sulfamethoxazole-trimethoprim, and streptomycin. These results also indicated that there was a correlation between antibiotic use and antimicrobial resistance.
In the study, there were four β-lactamase resistance genes detected. The β-lactamase-encoding genes prevalence was 34.3% in 67 E. coli isolates. β-lactamase resistance genes, such as blaCMY, blaSHV, blaCTX-M, and blaTEM were detected in nine non-pathogenic E. coli isolates. So non-pathogenic E. coli can serve as an antibiotic resistance reservoir and could possibly transfer genes to other pathogenic E. coli strains, which can pose a threat to mastitis management programs of farm (Hu et al., 2016). The rate of blaCTX-M, blaCMY, blaTEM, and blaSHV genes among E. coli was 1.5, 1.5, 10.4, and 20.9% in the study, respectively. The blaTEM and blaCMY genes were the most common, which is similar to several previous studies (Navajas-Benito et al., 2016; Gomi et al., 2017; Hinthong et al., 2017). The cephalosporins treatment in mastitis cattle also raised the proportion of blaTEM in milk samples at the period of withdrawal (p<0.05; Dong et al., 2021). The blaCTX-M, which was the most important ESBL-related gene, it was associated with the geographic area (Su et al., 2016). However, blaCTX-M was the most popular gene in Japan, United Kingdom, France, Netherlands, and Germany (Dahmen et al., 2013; Ohnishi et al., 2013; Timofte et al., 2014; Freitag et al., 2016; Santman-Berends et al., 2016).
Around 11.8% of E. coli stains showed resistance to tetracycline in the study. However, Su et al. (2016) reported that the tetracycline-resistance prevalence was 51%. Navajas-Benito et al. (2016) reported that antimicrobial resistance for tetracycline was detected in 19.2% of E. coli strains, which recovered from air and its surroundings in Spain. Antimicrobial resistance genes to tetracycline were tested in all the tetracycline-resistant isolates, and three tetracycline-resistant isolates harbored one tetracycline resistance gene tetB, which was the most frequent gene, and the studied E. coli did not possess tetA. However, Gomi et al. (2017) found that the prevalent of tetA was more than tetB in E. coli isolates. It was reported that one representative E. coli strain (No. JXLQYF114666) contained nine ARGs including aph(3'')-Ib, blaTEM-1B, blaCMY-2, aph(6)-Id, mdfA, sul2, tetB, catA2, and dfrA14, which result in resistance to seven important antibiotics classes (Liu et al., 2020). Moreover, the phenotype-genotype discrepancies on the tetracycline-resistant E. coli were observed in the study. However, resistance genotypes on tetracycline, gentamicin, kanamycin, and oxacillin correlated well with resistance phenotypes in E. coli and S. aureus (Gomi et al., 2017). Therefore, it was still necessary to fully account of testing phenotypic susceptibility for resistance (Zhao et al., 2015). Further research should be carried out to analyze the genetic characteristics on antibiotic resistance by whole-genome approach, which may explain the phenotype-genotype discrepancies observed for many strains.
Conclusion
In conclusion, the antibiotic resistance on E. coli isolated from raw milk in Northern China was assessed for the first time. Our data indicated that E. coli isolates were widely present in raw milk samples in Northern China. A total 20.9% of the tested E. coli possessed one or more virulence genes, which showed a potential pathogenicity. Escherichia coli strains exhibited different levels of antimicrobial resistance, except gentamicin. Ampicillin should not be a suitable treatment of dairy herds for mastitis by E. coli in Northern China. Majority of E. coli were multiple-antibiotic resistant and co-carried many virulence genes, and it may pose great potential risk to public health. The possibility of transferring and transmitting resistance genes, between non-pathogenic and pathogenic E. coli isolates, should be evaluated in further studies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
HL, LM, and LD designed and performed the research. YZ helped with the data analysis. JW gave advices to the researchers. NZ gave the opinions on the research design. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by China Agriculture Research System of MOF and MARA, The Agricultural Science and Technology Innovation Program (ASTIP-IAS12), The Scientific Research Project for Major Achievements of The Agricultural Science and Technology Innovation Program (CAAS-ZDXT2019004) and Project of Risk Assessment on Raw Milk (GJFP2019026).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.730656/full#supplementary-material
References
Awadallah, M. A., Ahmed, H. A., Merwad, A. M., and Selim, M. A. (2016). Occurrence, genotyping, Shiga toxin genes and associated risk factors of E. coli isolated from dairy farms, handlers and milk consumers. Vet. J. 217, 83–88. doi: 10.1016/j.tvjl.2016.09.014
Beutin, L., and Fach, P. (2015). “Detection of Shiga toxin-producing Escherichia coli from nonhuman sources and strain typing,” in Enterohemorrhagic Escherichia coli and Other Shiga Toxin-Producing E. coli (Washington, DC: American Society of Microbiology), 299–319.
Bhoomika,, Sanjay, S., Anil, P., and Eknath, G. N. (2016). Occurrence and characteristics of extended-spectrum β-lactamases producing Escherichia coli in foods of animal origin and human clinical samples in Chhattisgarh, India. Vet. World 9, 996–1000. doi: 10.14202/vetworld.2016.996-1000
Claeys, W. L., Cardoen, S., Daube, G., Block, J. D., and Herman, L. (2013). Raw or heated cow milk consumption: review of risks and benefits. Food Control 31, 251–262. doi: 10.1016/j.foodcont.2012.09.035
CLSI (2012). Performance Standards for Antimicrobial Susceptibility Testing. Document M100–S22. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI).
Cortés, C., Fuente, R., Blanco, J., Blanco, M., Blanco, J. E., Dhabi, G., et al. (2005). Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 110, 67–76. doi: 10.1016/j.vetmic.2005.06.009
Dahmen, S., Métayer, V., Gay, E., Madec, J. Y., and Haenni, M. (2013). Characterization of extended-spectrum beta-lactamase (esbl)-carrying plasmids and clones of enterobacteriaceae causing cattle mastitis in France. Vet. Microbiol. 162, 793–799. doi: 10.1016/j.vetmic.2012.10.015
Da Silva, G., and Mendonça, N. (2012). Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 3, 18–28. doi: 10.4161/viru.3.1.18382
Dong, L., Meng, L., Liu, H., Wu, H., Hu, H., Zheng, N., et al. (2021). Effect of therapeutic administration of β-lactam antibiotics on the bacterial community and antibiotic resistance patterns in milk. J. Dairy Sci. 104, 7018–7025. doi: 10.3168/jds.2020-20025
Douellou, T., Delannoy, S., Ganet, S., Mariani-Kurkdjian, P., Fach, P., Loukiadis, E., et al. (2016). Shiga toxin-producing Escherichia coli strains isolated from dairy products – genetic diversity and virulence gene profiles. Int. J. Food Microbiol. 232, 52–62. doi: 10.1016/j.ijfoodmicro.2016.04.032
EFSA (2015). Scientific opinion on the public health risks related to the consumption of raw drinking milk. EFSA J. 13:3940. doi: 10.2903/j.efsa.2015.4139
EFSA-ECDC (2012). Scientific report of EFSA and ECDC: the European Union summary report on trents and sources of zoonoses, agents and food-borne outbreaks in 2010. EFSA J. 10:2597. doi: 10.2903/j.efsa.2012.2669
Freitag, C., Michael, G. B., Kadlec, K., Hassel, M., and Schwarz, S. (2016). Detection of plasmid-borne extended-spectrum β-lactamase (ESBL) genes in Escherichia coli isolates from bovine mastitis. Vet. Microbiol. 200, 151–156. doi: 10.1016/j.vetmic.2016.08.010
Ghanbarpour, R., and Oswald, E. (2010). Phylogenetic distribution of virulence genes in Escherichia coli isolated from bovine mastitis in Iran. Res. Vet. Sci. 88, 6–10. doi: 10.1016/j.rvsc.2009.06.003
Gomez-Aldapa, C. A., Segovia-Cruz, J. A., Cerna-Cortes, J. F., Rangel-Vargas, E., Salas-Rangel, L. P., and Gutierrez-Alcantara, E. J. (2016). Prevalence and behavior of multidrug-resistant Shiga toxin-producing Escherichia coli, enteropathogenic E. coli and enterotoxigenic E. coli on coriander. Food Microbiol. 59, 97–103. doi: 10.1016/j.fm.2016.05.014
Gomi, R., Matsuda, T., Matsumura, Y., Yamamoto, M., Tanaka, M., Ichiyama, S., et al. (2017). Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in river water. Appl. Environ. Microbiol. 78:e02703-16. doi: 10.1128/AEM.02703-16
Gonggrijp, M. A., Santman-Berends, I., Heuvelink, A. E., Buter, G. J., and Lam, T. (2016). Prevalence and risk factors for extended-spectrum β-lactamase- and ampc-producing Escherichia coli in dairy farms. J. Dairy Sci. 99, 9001–9013. doi: 10.3168/jds.2016-11134
Hazen, T. H., Michalski, J., Luo, Q. W., Shetty, A. C., Daugherty, S. C., Fleckenstein, J. M., et al. (2017). Comparative genomics and transcriptomics of Escherichia coli isolates carrying virulence factors of both enteropathogenic and enterotoxigenic E. coli. Sci. Rep. 7:3513. doi: 10.1038/s41598-017-03489-z
Hernandes, R. T., Elias, W. P., Vieira, M. A., and Gomes, T. A. (2009). An overview of a typical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 297, 137–149. doi: 10.1111/j.1574-6968.2009.01664.x
Hinthong, W., Pumipuntu, N., Santajit, S., Kulpeanprasit, S., Buranasinsup, S., and Sookrung, N. (2017). Detection and drug resistance profile of Escherichia coli from subclinical mastitis cows and water supply in dairy farms in Saraburi Province, Thailand. PeerJ 5:e3431. doi: 10.7717/peerj.3431
Hu, Y., Yang, X., Li, J., Lv, N., Liu, F., Wu, J., et al. (2016). The transfer network of bacterial mobile resistome connecting animal and human microbiome. Appl. Environ. Microbiol. 82, 6672–6681. doi: 10.1128/AEM.01802-16
Islam, M. A., Kabir, S. M. L., and Seel, S. K. (2016). Molecular detection and characterization of Escherichia coli isolated from raw milk sold in different markets of Bangladesh. Bangladesh J. Vet. Med. 14, 271–275. doi: 10.3329/bjvm.v14i2.31408
Jayarao, B. M., Donaldson, S. C., Straley, B. A., Sawant, A. A., Hegde, N. V., and Brown, J. L. (2006). A survey of foodborne pathogens in bulk tank milk and raw milk consumption among farm families in Pennsylvania. J. Dairy Sci. 89, 2451–2458. doi: 10.3168/jds.S0022-0302(06)72318-9
Jayarao, B. M., and Henning, D. R. (2001). Prevalence of foodborne pathogens in bulk tank milk. J. Dairy Sci. 84, 2157–2162. doi: 10.3168/jds.S0022-0302(01)74661-9
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Lambertini, E., Karns, J. S., Kessel, J., Cao, H., and Pradhan, A. K. (2015). Dynamics of Escherichia coli virulence factors in dairy herds and farm environments in a longitudinal study in the United States. Appl. Environ. Microbiol. 81, 4477–4488. doi: 10.1128/AEM.00465-15
Lan, X. Y., Zhao, S. G., Zheng, N., Li, S. L., Zhang, Y. D., Liu, H. M., et al. (2017). Short communication: microbiological quality of raw milk of raw cow milk and its association with herd management practices in Northern China. J. Dairy Sci. 100, 4294–4299. doi: 10.3168/jds.2016-11631
Lippolis, J. D., Holman, D. B., Brunelle, B. W., Thacker, T. C., Bearson, B. L., and Reinhardt, T. A. (2017). Genomic and transcriptomic analysis of Escherichia coli strains associated with persistent and transient bovine mastitis and the role of colanic acid. Infect. Immun. 86:e00566-17. doi: 10.1128/IAI.00566-17
Liu, H. M., Li, S. L., Meng, L., Dong, L., Zhao, S. G., Lan, X. Y., et al. (2017). Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J. Dairy Sci. 100, 8796–8803. doi: 10.3168/jds.2017-13370
Liu, J., Zhu, Y., Jay-Russell, M., Lemay, D. G., and Mills, D. A. (2020). Reservoirs of antimicrobial resistance genes in retail raw milk. Microbiome 8:99. doi: 10.1186/s40168-020-00861-6
McConnel, C. S., Stenkamp-Strahm, C. M., Rao, S., Linke, L. M., Magnuson, R. J., and Hyatt, D. R. (2016). Antimicrobial resistance profiles in Escherichia coli O157 isolated from Northern Colorado dairies. J. Food Prot. 79, 484–487. doi: 10.4315/0362-028X.JFP-15-321
Medeiros, P., Bolick, D., Roche, J. K., Noronha, F., Pinheiro, C., Kolling, G., et al. (2013). The micronutrient zinc inhibits EAEC strain 042 adherence, biofilm formation, virulence gene expression, and epithelial cytokine responses benefiting the infected host. Virulence 4, 624–633. doi: 10.4161/viru.26120
Nam, H. M., Lim, S. K., Kang, H. M., Kim, J. M., Moon, J. S., and Jang, K. C. (2009). Prevalence and antimicrobial susceptibility of gram-negative bacteria isolated from bovine mastitis between 2003 and 2008 in Korea. J. Dairy Sci. 92, 2020–2026. doi: 10.3168/jds.2008-1739
Navajas-Benito, E. V., Alonso, C. A., Sanz, S., Olarte, C., Martínez-Olarte, R., and Hidalgo-Sanz, S. (2016). Molecular characterization of antibiotic resistance in Escherichia coli strains from a dairy cattle farm and its surroundings. J. Sci. Food Agric. 97, 363–365. doi: 10.1002/jsfa.7709
Ntuli, V., Njage, P. M. K., and Buys, E. M. (2016). Characterization of Escherichia coli and other Enterobacteriaceae in producer-distributor bulk milk. J. Dairy Sci. 99, 9534–9549. doi: 10.3168/jds.2016-11403
Ohnishi, M., Okatani, A. T., Harada, K., Sawada, T., and Takahashi, T. (2013). Genetic characteristics of CTX-M-type extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae involved in mastitis cases on Japanese dairy farms, 2007 to 2011. J. Clin. Microbiol. 51, 3117–3122. doi: 10.1128/JCM.00920-13
Ombarak, R. A., Hinenoya, A., Awasthi, S. P., Iguchi, A., Shima, A., Elbagory, A. R. M., et al. (2016). Prevalence and pathogenic potential of Escherichia coli isolates from raw milk and raw milk cheese in Egypt. Int. J. Food. Microbial. 221, 69–76. doi: 10.1016/j.ijfoodmicro.2016.01.009
Pradel, N., Bertin, Y., Martin, C., and Livrelli, V. (2008). Molecular analysis of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France. Appl. Environ. Microbiol. 74, 2118–2128. doi: 10.1128/AEM.02688-07
Ribeiro, L. F., Barbosa, M. M. C., Pinto, F. R., Maluta, R. P., Oliveira, M. C., de Souza, V., et al. (2016). Antimicrobial resistance and virulence factors of Escherichia coli in cheese made from unpasteurized milk in three cities in Brazil. Foodborne Pathog. Dis. 13, 469–476. doi: 10.1089/fpd.2015.2106
Radostits, O. M., Gay, C. C., and Hinchcliff, K. W. (2007). Veterinary Medicine— A textbook of the diseases of cattle, horses, sheep, pigs, and goats. 10.ed. Philadelphia: Saunders. 673–762.
Ruegg, P. L., Oliveira, L., Jin, W., and Okwumabua, O. (2014). Phenotypic antimicrobial susceptibility and occurrence of selected resistance genes in gram-positive mastitis pathogens isolated from Wisconsin dairy cows. J. Dairy Sci. 98, 4521–4534. doi: 10.3168/jds.2014-9137
Rugeles, L. C., Bai, J., Martinez, A. J., Vanegas, M. C., and Gomez-Duarte, O. G. (2010). Molecular characterization of diarrheagenic Escherichia coli strains from stools samples and food products in Colombia. Int. J. Food Microbiol. 138, 282–286. doi: 10.1016/j.ijfoodmicro.2010.01.034
Santman-Berends, I., Gonggrijp, M. A., Hage, J. J., Heuvelink, A. E., Velthuis, A., and Lam, T. (2016). Prevalence and risk factors for extended-spectrum beta-lactamase or AMPC-producing Escherichia coli in organic dairy herds in the Netherlands. J. Dairy Sci. 562, 120–128. doi: 10.3168/jds.2016-11839
Sharma, S., Aarif, K., Dahiya, D. K., Jain, J., and Sharma, V. (2015). Prevalence, identification and drug resistance pattern of Staphylococcus aureus and Escherichia coli isolated from raw milk samples of Jaipur city of Rajasthan. J. Pure Appl. Microbiol. 9, 341–348.
Su, Y. C., Yu, C. Y., Tsai, Y. L., Wang, S. H., Lee, C. h., and Chu, C. (2016). Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli from the milk of cows with clinical mastitis in Southern Taiwan. J. Microbiol. Immunol. Infect. 49, 892–901. doi: 10.1016/j.jmii.2014.10.003
Suojala, L., Pohjanvirta, T., Simojoki, H., Myllyniemi, A. L., Pitkala, A., Pelkonen, S., et al. (2011). Phylogeny, virulence factors and antimicrobial susceptibility of Escherichia coli isolated in clinical bovine mastitis. Vet. Microbiol. 147, 383–388. doi: 10.1016/j.vetmic.2010.07.011
Tabaran, A., Mihaiu, M., Tabaran, F., Colobatiu, L., Reget, O., Borzan, M. M., et al. (2017). First study on characterization of virulence and antibiotic resistance genes in verotoxigenic and enterotoxigenic E. coli isolated from raw milk and unpasteurized traditional cheeses in Romania. Folia Microbiol. 62, 145–150. doi: 10.1007/s12223-016-0481-8
Tark, D. S., Moon, D. C., Kang, H. Y., Kim, S. R., Nam, H. M., Lee, H. S., et al. (2016). Antimicrobial susceptibility and characterization of extended-spectrum β-lactamases in Escherichia coli isolated from bovine mastitic milk in South Korea from 2012 to 2015. J. Dairy Sci. 100, 3463–3469. doi: 10.3168/jds.2016-12276
Timofte, D., Maciuca, I. E., Evans, N. J., Williams, H., Wattret, A., and Fick, J. C. (2014). Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob. Agents Chemother. 58, 789–794. doi: 10.1128/AAC.00752-13
Trevisani, M., Mancusi, R., Donne, G. D., Bacci, C., Bassi, L., and Bonardi, S. (2014). Detection of Shiga toxin (Stx)-producing Escherichia coli (STEC) in bovine dairy herds in northern Italy. Int. J. Food Microbiol. 184, 45–49. doi: 10.1016/j.ijfoodmicro.2013.12.033
Van Kessel, J. A. S., Karns, J. S., Lombard, J. E., and Kopral, A. C. A. (2011). Prevalence of Salmonella enterica, listeria monocytogenes, and Escherichia coli virulence factors in bulk tank milk and in-line filters from US dairies. J. Food Prot. 74, 759–768. doi: 10.4315/0362-028X.JFP-10-423
Wang, D., Zhang, L. M., Zhou, X. Z., He, Y. L., Yong, C. H., Shen, M. L., et al. (2016). Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J. Dairy Sci. 99, 9560–9569. doi: 10.3168/jds.2016-11625
Zhang, S. H., Wu, Q. P., Zhang, J. M., Lai, Z. P., and Zhu, X. M. (2016). Prevalence, genetic diversity, and antibiotic resistance of enterotoxigenic Escherichia coli in retail ready to eat foods in China. China. Food Control. 68, 236–243. doi: 10.1016/j.foodcont.2016.03.051
Keywords: Escherichia coli, virulence, antimicrobial resistance, raw milk, Northern China
Citation: Liu H, Meng L, Dong L, Zhang Y, Wang J and Zheng N (2021) Prevalence, Antimicrobial Susceptibility, and Molecular Characterization of Escherichia coli Isolated From Raw Milk in Dairy Herds in Northern China. Front. Microbiol. 12:730656. doi: 10.3389/fmicb.2021.730656
Edited by:
Fereidoun Forghani, IEH Laboratories and Consulting Group, United StatesReviewed by:
Jinxin Liu, Nanjing Agricultural University, ChinaKai Deng, IEH Laboratories and Consulting Group, United States
Copyright © 2021 Liu, Meng, Dong, Zhang, Wang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Zheng, zhengnan_1980@126.com
†These authors have contributed equally to this work and share first authorship
 Huimin Liu
Huimin Liu Lu Meng1,2†
Lu Meng1,2† Lei Dong
Lei Dong Jiaqi Wang
Jiaqi Wang