Protection against Oxidative Stress and Metabolic Alterations by Synthetic Peptides Derived from Erythrina edulis Seed Protein
Abstract
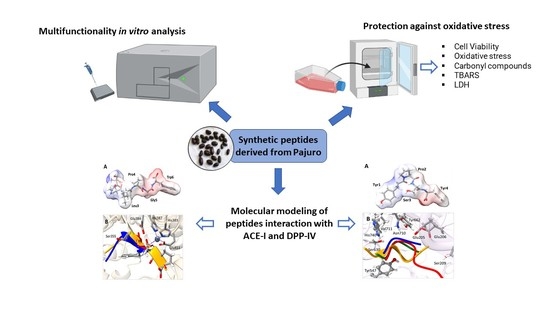
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Obtention of Synthetic Peptides from Pajuro
2.3. Antioxidant Activity of Synthetic Pajuro-Derived Peptides
2.4. Angiotensin-Converting Enzyme (ACE) Inhibitory Activity
2.5. In Vitro Anti-Diabetic Activity
2.6. Pancreatic Lipase Inhibitory Activity
2.7. Protective Effects of Synthetic Pajuro-Derived Peptides in SH-SY5Y Cells
2.7.1. Effects on Cell Viability
2.7.2. Protective Effects against Oxidative Stress Induced by FeSO4
2.7.3. Determination of Thiobarbituric Acid Reaction Substances (TBARS)
2.7.4. Determination of Lactate Dehydrogenase (LDH) Activity
2.7.5. Determination of Carbonyl Compounds
2.8. Molecular Modeling of Pajuro-Derived Peptides Interaction with ACE-I and DPP-IV
2.9. Statistical Analysis
3. Results and Discussion
3.1. In Vitro Multifunctionality of Pajuro-Derived Peptides
3.2. Neuroprotective Effects of Pajuro-Derived Peptides
3.3. Molecular Modeling of Pajuro-Derived Peptides Interaction with ACE-I and DPP-IV
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). World Health Statistics 2018: Monitoring Health for the Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Olivieri, C. The current state of heart disease: Statins, cholesterol, fat and sugar. Int. J. Evid.-Based Healthc. 2019, 17, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Ghaedi, E.; Mohammadi, M.; Mohammadi, H.; Ramezani-Jolfaie, N.; Malekzadeh, J.; Hosseinzadeh, M.; Salehi-Abargouei, A. Effects of a paleolithic diet on cardiovascular disease risk factors: A systematic review and meta-analysis of randomized controlled trials. Adv. Nutr. 2019, 10, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Koch, W. Dietary polyphenols-important non-nutrients in the prevention of chronic noncommunicable diseases. A systematic review. Nutrients 2019, 11, 1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waris, G.; Ahsan, H. Reactive oxygen species: Role in the development of cancer and various chronic conditions. J. Carcinog. 2006, 5, 14. [Google Scholar] [CrossRef]
- Kaneto, H.; Katakami, N.; Matsuhisa, M.; Matsuoka, T. Role of reactive oxygen species in the progression of type 2 diabetes and atherosclerosis. Mediators Inflamm. 2010, 2010, 453892. [Google Scholar] [CrossRef] [Green Version]
- Ciau-Solis, N.A.; Acevedo-Fernandez, J.J.; Betancur-Ancona, D. In vitro renin-angiotensin system inhibition and in vivo antihypertensive activity of peptide fractions from lima bean (Phaseolus lunatus L.). J. Sci. Food Agric. 2018, 98, 781–786. [Google Scholar] [CrossRef]
- Koch, W.; Baj, T.; Kukula-Koch, W.; Marzec, Z. Dietary intake of specific phenolic compounds and their effect on the antioxidant activity of daily food rations. Open Chem. 2015, 13, 869–876. [Google Scholar] [CrossRef]
- Daliri, E.B.; Oh, D.H.; Lee, B.H. Bioactive peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef]
- Lammi, C.; Aiello, G.; Boschin, G.; Arnoldi, A. Multifunctional peptides for the prevention of cardiovascular disease: A new concept in the area of bioactive food-derived peptides. J. Funct. Foods 2019, 55, 135–145. [Google Scholar] [CrossRef]
- de Fatima Garcia, B.; de Barros, M.; de Souza Rocha, T. Bioactive peptides from beans with the potential to decrease the risk of developing noncommunicable chronic diseases. Crit. Rev. Food Sci. Nutr. 2021, 61, 2003–2021. [Google Scholar] [CrossRef]
- Matemu, A.; Nakamura, S.; Katayama, S. Health benefits of antioxidative peptides derived from legume proteins with a high amino acid score. Antioxidants 2021, 10, 316. [Google Scholar] [CrossRef] [PubMed]
- Mojica, L.; Luna-Vital, D.; González de Mejía, E. Characterization of peptides from common bean protein isolates and their potential to inhibit markers of type-2 diabetes, hypertension and oxidative stress. J. Sci. Food Agric. 2017, 97, 2401–2410. [Google Scholar] [CrossRef] [PubMed]
- Jakubczyk, A.; Karaś, M.; Złotek, U.; Szymanowska, U. Identification of potential inhibitory peptides of enzymes involved in the metabolic syndrome obtained by simulated gastrointestinal digestion of fermented bean (Phaseolus vulgaris L.) seeds. Food Res. Int. 2017, 100, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Tomé, S.; Hernández-Ledesma, B. Current state of art after twenty years of the discovery of bioactive peptide lunasin. Food Res. Int. 2019, 116, 71–78. [Google Scholar] [CrossRef]
- Arango Bedoya, O.; Bolaños Patiño, V.; Ricaurte García, D.; Caicedo, M.; Guerrero, Y. Obtaining a protein extract from chachafruto flour (Erythrina edulis). Rev. Univ. Salud 2012, 14, 161–167. [Google Scholar]
- Sánchez Chero, M.J.; Sánchez Chero, J.A.; Miranda Zamora, W. Technify and conserve the bioactive components of Pashul (Erythrina edulis) for human consumption. UCV HACER Rev. Inv. Cult. 2019, 8, 11–17. [Google Scholar]
- Intiquilla, I.; Jiménez-Aliaga, K.; Zavaleta, A.I.; Arnao, I.; Peña, C.; Chávez-Hidalgo, E.L.; Hernández-Ledesma, B. Erythrina edulis (pajuro) seed protein: A new source of antioxidant peptides. Nat. Prod. Commun. 2016, 11, 781–786. [Google Scholar] [CrossRef] [Green Version]
- Re, R.; Pellergrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Hernández-Ledesma, B.; Dávalos, A.; Bartolomé, B.; Amigo, L. Preparation of antioxidant enzymatic hydrolysates from α-lactalbumin and β-lactoglobulin. Identification of active peptides by HPLC-MS/MS. J. Agric. Food Chem. 2005, 53, 588–593. [Google Scholar] [CrossRef]
- Hayakari, M.; Kondo, Y.; Izumi, H. A rapid and simple spectrophotometric assay of angiotensin-converting enzyme. Anal. Biochem. 1978, 84, 361–369. [Google Scholar] [CrossRef]
- Subramanian, R.; Asmawi, M.Z.; Sadikun, A. In vitro α-glucosidase and α-amylase enzyme inhibitory effects of Andrographis paniculata extract and andrographolide. Acta Biochim. Pol. 2008, 55, 391–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar Singla, R.; Singh, R.; Kumar Dubey, A. Important aspects of post-prandial antidiabetic drug, acarbose. Curr. Top. Med. Chem. 2016, 16, 2625–2633. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Lucius, A.; Meyer, T.; De Mejia, E. Cultivar evaluation and effect of fermentation on antioxidant capacity and in vitro inhibition of alpha-amylase and alpha-glucosidase by highbush blueberry (Vaccinium corombosum). J. Agric. Food Chem. 2011, 59, 8923–8930. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yu, Z.; Zhu, H.; Zhang, W.; Chen, Y. In vitro alpha-glucosidase inhibitory activity of isolated fractions from water extract of Qingzhuan dark tea. BMC Complement. Altern. Med. 2016, 16, 378. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Rathi, P.; Gupta, R. Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal. Biochem. 2002, 311, 98–99. [Google Scholar] [CrossRef]
- Dolenc, A.; Govedarica, B.; Dreu, R.; Kocbek, P.; Srčič, S.; Kristl, J. Nanosized particles of orlistat with enhanced in vitro dissolution rate and lipase inhibition. Int. J. Pharm. 2010, 396, 149–155. [Google Scholar] [CrossRef]
- Palma-Albino, C.; Intiquilla, A.; Jiménez-Aliaga, K.; Rodríguez-Arana, N.; Solano, E.; Flores, E.; Zavaleta, A.I.; Izaguirre, V.; Hernández-Ledesma, B. Albumin from Erythrina edulis (Pajuro) as a promising source of multifunctional peptides. Antioxidants 2021, 10, 1722. [Google Scholar] [CrossRef]
- LeBel, C.P.; Ischiropoulos, H.; Bondy, S.C. Evaluation of the probe 2′,7′-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem. Res. Toxicol. 1992, 5, 227–231. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Aliaga, K.; Bermejo-Besco, P.; Benedi, J.; Martin-Aragon, S. Quercetin and rutin exhibit antiamyloidogenic and fibril-disaggregating effects in vitro and potent antioxidant activity in APPswe cells. Life Sci. 2022, 89, 939–945. [Google Scholar] [CrossRef]
- Thoma, R.; Löffler, B.; Stihle, M.; Huber, W.; Ruf, A.; Hennig, M. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure 2003, 11, 947–959. [Google Scholar] [CrossRef] [Green Version]
- Natesh, R.; Schwager, S.L.; Evans, H.R.; Sturrock, E.D.; Acharya, K.R. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry 2004, 43, 8718–8724. [Google Scholar] [CrossRef] [PubMed]
- Land, H.; Humble, M.S. YASARA: A tool to obtain structural guidance in biocatalytic investigations. Methods Mol. Biol. 2018, 1685, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006, 25, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Krüger, D.M.; Gohlke, H. DrugScorePPI webserver: Fast and accurate in silico alanine scanning for scoring protein-protein interactions. Nucleic Acids Res. 2010, 38, W480–W486. [Google Scholar] [CrossRef]
- Konagurthu, A.S.; Whisstock, J.C.; Stuckey, P.J.; Lesk, A.M. MUSTANG: A multiple structural alignment algorithm. Proteins 2006, 64, 559–574. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Amigo, L.; Martínez-Maqueda, D.; Hernández-Ledesma, B. In silico and in vitro analysis of multifunctionality of animal food-derived peptides. Foods 2020, 9, 991. [Google Scholar] [CrossRef]
- Intiquilla, A.; Jimenez-Aliaga, K.; Guzman, F.; Alvarez, C.A.; Zavaleta, A.I.; Izaguirre, V.; Hernandez-Ledesma, B. Novel antioxidant peptides obtained by alcalase hydrolysis of Erythrina edulis (pajuro) protein. J. Sci. Food Agric. 2019, 99, 2420–2427. [Google Scholar] [CrossRef]
- Hernandez-Ledesma, B.; Miralles, B.; Amigo, L.; Ramos, M.; Recio, I. Identification of antioxidant and ACE-inhibitory peptides in fermented milk. J. Sci. Food Agric. 2005, 85, 1041–1048. [Google Scholar] [CrossRef]
- Samaranayaka, A.G.P.; Li-Chan, E.C.Y. Food-derived peptidic antioxidant: A review of their production, assessment, and potential applications. J. Funct. Foods 2011, 3, 229–254. [Google Scholar] [CrossRef]
- Cheung, H.-S.; Wang, F.-L.; Ondetti, M.A.; Sabo, E.F.; Cushman, D.W. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence. J. Biol. Chem. 1980, 255, 401–407. [Google Scholar] [CrossRef]
- Ren, J.; Chen, S.; Li, C.; Gu, Z.; Cheng, L.; Hong, Y.; Li, Z. A two-stage modification method using 1,4-α-glucan branching enzyme lowers the in vitro digestibility of corn starch. Food Chem. 2020, 305, 25441. [Google Scholar] [CrossRef] [PubMed]
- Siow, H.-L.; Lim, T.S.; Gan, C.-Y. Development of a workflow for screening and identification of a-amylase inhibitory peptides from food source using an integrated Bioinformatics-phage display approach: Case study—Cumin seed. Food Chem. 2017, 214, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Elam, E.; Feng, J.; Lv, Y.-M.; Ni, Z.-J.; Sun, P.; Thakur, K.; Zhang, J.-G.; Ma, Y.-L.; Wei, Z.-J. Recent advances on bioactive food derived anti-diabetic hydrolysates and peptides from natural resources. J. Funct. Foods 2021, 86, 104674. [Google Scholar] [CrossRef]
- Vilcacundo, R.; Martínez-Villaluenga, C.; Hernández-Ledesma, B. Release of dipeptidyl peptidase IV, α-amylase and α-glucosidase inhibitory peptides from quinoa (Chenopodium quinoa Willd.) during in vitro simulated gastrointestinal digestion. J. Funct. Foods 2017, 35, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Ngoh, Y.Y.; Gan, C.Y. Enzyme-assisted extraction and identification of antioxidative and alpha-amylase inhibitory peptides from Pinto beans (Phaseolus vulgaris cv. Pinto). Food Chem. 2016, 190, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Bu, Y.G.; Zhao, M.L.; Tao, R.; Li, Y. Studies on antioxidant and α-glucosidase inhibitory constituents of chinese toon bud (Toona sinensis). J. Funct. Foods 2020, 73, 104108. [Google Scholar] [CrossRef]
- Tian, J.L.; Si, X.; Wang, Y.H.; Gong, E.S.; Xie, X.; Zhang, Y. Bioactive flavonoids from Rubus corchorifolius inhibit α-glucosidase and α-amylase to improve postprandial hyperglycemia. Food Chem. 2020, 341, 128149. [Google Scholar] [CrossRef]
- Roskar, I.; Molek, P.; Vodnik, M.; Stempelj, M.; Strukelj, B.; Lunder, M. Peptide modulators of alpha-glucosidase. J. Diabetes Investig. 2015, 6, 625–631. [Google Scholar] [CrossRef]
- You, Q.; Chen, F.; Wang, X.; Jiang, Y.; Lin, S. Anti-diabetic activities of phenolic compounds in muscadine against alpha-glucosidase and pancreatic lipase. Food Sci. Technol. 2012, 46, 164–168. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Inhibition of dipeptidyl peptidase (DPP)-IV and a-glucosidase activities by pepsin-treated whey proteins. J. Agric. Food Chem. 2013, 61, 7500–7506. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Liang, K.; Jin, Y.; Zhang, M.; Chen, Y.; Wu, H.; Lai, F. Identification and characterization of two novel a-glucosidase inhibitory oligopeptides from hemp (Cannabis sativa L.) seed protein. J. Funct. Foods 2016, 26, 439–450. [Google Scholar] [CrossRef]
- Oseguera-Toledo, M.E.; Gonzalez de Mejia, E.; Sivaguru, M.; Amaya-Llano, S.L. Common bean (Phaseolus vulgaris L.) protein-derived peptides increased insulin secretion, inhibited lipid accumulation, increased glucose uptake and reduced the phosphatase and tensin homologue activation in vitro. J. Funct. Foods 2016, 27, 160–177. [Google Scholar] [CrossRef]
- Feng, J.; Ma, Y.L.; Sun, P.; Thakur, K.; Wang, S.Y.; Zhang, J.G. Purification and characterisation of α-glucosidase inhibitory peptides from defatted camellia seed cake. Int. J. Food Sci. Technol. 2021, 56, 138–147. [Google Scholar] [CrossRef]
- Bharatham, K.; Bharatham, N.; Park, K.H.; Lee, K.W. Binding mode analyses and pharmacophore model development for sulfonamide chalcone derivatives, a new class of α-glucosidase inhibitors. J. Mol. Graph. Model. 2008, 26, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Matteucci, E.; Giampietro, O. Dipeptidyl peptidase-4 (CD26): Knowing the function before inhibiting the enzyme. Curr. Med. Chem. 2009, 16, 2943–2951. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M. Synthesis, evaluation and molecular docking of thiazolopyrimidine derivatives as dipeptidyl peptidase iv inhibitors. Chem. Biol. Drug Des. 2020, 80, 918–928. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Features of dipeptidyl peptidase IV (DPPIV) inhibitory peptides from dietary proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela Zamudio, F.; Segura Campos, M.R. Amaranth, quinoa and chia bioactive peptides: A comprehensive review on three ancient grains and their potential role in management and prevention of Type 2 diabetes. Crit. Rev. Food Sci. Nutr. 2022, 62, 2707–2721. [Google Scholar] [CrossRef]
- Velarde-Salcedo, A.J.; Barrera-Pacheco, A.; Lara-González, S.; Montero-Morán, G.M.; Díaz-Gois, A.; González de Mejia, E.; Barba de la Rosa, A.P. In vitro inhibition of dipeptidyl peptidase IV by peptides derived from the hydrolysis of amaranth (Amaranthus hypochondriacus L.) proteins. Food Chem. 2013, 136, 758–764. [Google Scholar] [CrossRef]
- Lunder, M.; Bratkovic, T.; Kreft, S.; Strukelj, B. Peptide inhibitor of pancreatic lipase selected by phage display using different elution strategies. J. Lipid Res. 2005, 46, 1512–1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunagariya, N.A.B.; Patel, N.K.; Jagtap, S.C.; Bhutani, K.K. Inhibitors of pancreatic lipase: State of the art and clinical perspectives. EXCLI J. 2014, 13, 897–921. [Google Scholar]
- Awosika, T.O.; Aluko, R.E. Inhibition of the in vitro activities of a-amylase, a-glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. Int. J. Food Sci. Technol. 2019, 54, 2021–2034. [Google Scholar] [CrossRef]
- Moreno Valdespino, C.; Gonzalez de Mejia, E.; Mojica, L.; Luna-Vital, D.; Camacho, R. Bioactive peptides from black bean proteins play a potential role in the prevention of adipogenesis. Curr. Dev. Nutr. 2019, 3, nzz031.P06-119-19. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.J.; Lee, J.Y.; Seo, S.R.; Chung, K.C. Overexpression of DSCR1 blocks zincinduced neuronal cell death through the formation of nuclear aggregates. Mol. Cell Neurosci. 2007, 35, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.; Kantham, S.; Rao, V.M.; Palanivelu, M.K.; Pham, H.L.; Shaw, P.N.; McGeary, R.P.; Ross, B.P. Metal chelation, radical scavenging and inhibition of Aβ42 fibrillation by food constituents in relation to Alzheimer’s disease. Food Chem. 2016, 199, 185–194. [Google Scholar] [CrossRef] [Green Version]
- Carrera, I.; Cacabelos, R. Current drugs and potential future neuroprotective compounds for Parkinson’s disease. Curr. Neuropharmacol. 2019, 17, 295–306. [Google Scholar] [CrossRef]
- Feng, L.; Peng, F.; Wang, X.; Li, M.; Lei, H.; Xu, H. Identification and characterization of antioxidative peptides derived from simulated in vitro gastrointestinal digestion of walnut meal proteins. Food Res. Int. 2019, 116, 518–526. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hur, S.J. Neuroprotective effects of different molecular weight peptide fractions obtained from beef by hydrolysis with commercial enzymes in SH-SY5Y cells. Food Res. Int. 2019, 121, 176–184. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hur, S.J. Protective effect of a 3 kDa peptide obtained from beef myofibrillar protein using alkaline-AK on neuronal cells. Neurochem. Int. 2019, 129, 104459. [Google Scholar] [CrossRef]
- Zhang, Q.; Tong, X.; Li, Y.; Wang, H.; Wang, Z.; Qi, B.; Sui, X.; Jiang, L. Purification and characterization of antioxidant peptides from alcalase-hydrolyzed soybean (Glycine max L.) hydrolysate and their cytoprotective effects in human intestinal Caco-2 cells. J. Agric. Food Chem. 2019, 67, 5772–5781. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shao, B.; Yu, H.; Xu, F.; Wang, P.; Yu, K.; Han, Y.; Song, M.; Li, Y.; Cao, Z. Neuroprotective role of hyperforin on aluminum maltolate-induced oxidative damage and apoptosis in PC12 cells and SH-SY5Y cells. Chem. Biol. Interact. 2019, 299, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Cheng, S.; Dong, Y.; Ju, H. Intracellular antioxidant activity and apoptosis inhibition capacity of PEF-treated KDHCH in HepG2 cells. Food Res. Int. 2019, 121, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.R.; Khalil, S.R.; Zaglool, A.W.; Hendam, B.M.; Moustafa, A.A.; Cocco, R.; Di Cerbo, A.; Alagawany, M. Thiacloprid induced developmental neurotoxicity via ROS-oxidative injury and inflammation in chicken embryo: The possible attenuating role of chicoric and rosmarinic acids. Biology 2021, 10, 1100. [Google Scholar] [CrossRef]
- Mirzaei, M.; Mirdamadi, S.; Safavi, M. Antioxidant activity and protective effects of Saccharomyces cerevisiae peptide fractions against H2O2-induced oxidative stress in Caco-2 cells. J. Food Meas. Charact. 2019, 13, 2654–2662. [Google Scholar] [CrossRef]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, G.E.M.; Govender, T.; Naicker, T.; Kruger, H.G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. 2021, 224, 113705. [Google Scholar] [CrossRef]
- Hayes, M. Bioactive peptides in preventative healthcare: An overview of bioactivities and suggested methods to assess potential applications. Curr. Pharm. Des. 2021, 27, 1332–1341. [Google Scholar] [CrossRef]
- Martini, S.; Cattivelli, A.; Conte, A.; Tagliazucchi, D. Application of a combined peptidomics and in silico approach for the identification of novel dipeptidyl peptidase-IV-inhibitory peptides in in vitro digested pinto bean protein extract. Curr. Issues Mol. Biol. 2021, 44, 11. [Google Scholar] [CrossRef]
- Mirabito Colafella, K.M.; Bovée, D.M.; Danser, A.H.J. The renin-angiotensin-aldosterone system and its therapeutic targets. Exp. Eye Res. 2019, 186, 107680. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014, 165, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Evaluation of the potential of dietary proteins as precursors of dipeptidyl peptidase (DPP)-IV inhibitors by an in silico approach. J. Funct. Foods 2012, 4, 403–422. [Google Scholar] [CrossRef]
- Kalyan, G.; Junghare, V.; Khan, M.F.; Pal, S.; Bhattacharya, S.; Guha, S.; Majumder, K.; Chakrabarty, S.; Hazra, S. Anti-hypertensive peptide predictor: A machine learning-empowered web server for prediction of food-derived peptides with potential angiotensin-converting enzyme-I inhibitory activity. J. Agric. Food Chem. 2021, 69, 14995–15004. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vaquero, M.; Mora, L.; Hayes, M. In vitro and in silico approaches to generating and identifying angiotensin-converting enzyme I inhibitory peptides from green macroalga Ulva lactuca. Mar. Drugs 2019, 17, 204. [Google Scholar] [CrossRef] [PubMed]
- Gangopadhyay, N.; Wynne, K.; O’Connor, P.; Gallagher, E.; Brunton, N.P.; Rai, D.K.; Hayes, M. In silico and in vitro analyses of the angiotensin-I converting enzyme inhibitory activity of hydrolysates generated from crude barley (Hordeum vulgare) protein concentrates. Food Chem. 2016, 203, 367–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, H.; Liao, W.; Wu, J. Molecular interactions, bioavailability, and cellular mechanisms of angiotensin-converting enzyme inhibitory peptides. J. Food Biochem. 2019, 43, e12572. [Google Scholar] [CrossRef] [Green Version]
- Jimsheena, V.K.; Gowda, L.R. Arachin derived peptides as selective angiotensin I-converting enzyme (ACE) inhibitors: Structure-activity relationship. Peptides 2010, 31, 1165–1176. [Google Scholar] [CrossRef]
- Pina, A.S.; Roque, A.C. Studies on the molecular recognition between bioactive peptides and angiotensin-converting enzyme. J. Mol. Recognit. 2009, 22, 162–168. [Google Scholar] [CrossRef]
- Masuyer, G.; Schwager, S.L.; Sturrock, E.D.; Isaac, R.E.; Acharya, K.R. Molecular recognition and regulation of human angiotensin-I converting enzyme (ACE) activity by natural inhibitory peptides. Sci. Rep. 2012, 2, 717. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Inhibition of dipeptidyl peptidase IV (DPP-IV) by proline containing peptides. J. Funct. Foods 2013, 5, 1909–1917. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, H.; Aoyagi, T.; Ogawa, K.; Naganawa, H.; Hamada, M.; Takeuchi, T. Diprotins A and B, inhibitors of dipeptidyl aminopeptidase IV, produced by bacteria. J. Antibiot. 1984, 37, 422–425. [Google Scholar] [CrossRef] [Green Version]
- Nabeno, M.; Akahoshi, F.; Kishida, H.; Miyaguchi, I.; Tanaka, Y.; Ishii, S.; Kadowaki, T. A comparative study of the binding modes of recently launched dipeptidyl peptidase IV inhibitors in the active site. Biochem. Biophys. Res. Commun. 2013, 434, 191–196. [Google Scholar] [CrossRef] [PubMed]




| Peptide | ABTS (μmol TE/μmol Péptide) | ORAC (μmol TE/μmol Peptide) | ACE Inhibition (IC50 = μM) | α-Amylase Inhibition a (%) | α-Glucosidase Inhibition a (%) | DPP-IV Inhibition (IC50 = μM) | Pancreatic lipase Inhibition a (%) |
|---|---|---|---|---|---|---|---|
| Control | --- | --- | 6.81 ± 0.04 | 75.05 ± 1.55 | 62.36 ± 0.54 | 6.80 ± 0.03 | 94.62 ± 0.90 |
| GPPW | 0.26 ± 0.01 | 2.96 ± 0.38 | n.d. | n.d | n.d. | n.d. | n.d |
| TWVV | 0.11 ± 0.01 | 2.27 ± 0.07 | n.d. | 24.30 ± 0.01 | n.d. | n.d. | n.d |
| YPSY | 1.13 ± 0.06 | 3.26 ± 0.21 | 115.60 ± 0.26 | n.d. | n.d. | 32.60 ± 1.60 | 15.56 ± 1.35 |
| AALWE | 0.80 ± 0.50 | 1.05 ± 0.20 | n.d. | n.d. | n.d. | 130.60 ± 2.34 | 18.19 ± 0.98 |
| YYLTR | 0.40 ± 0.01 * | 1.04 ± 0.07 * | n.d. | n.d | n.d. | n.d. | n.d |
| CCGDYY | 1.18 ± 0.03 * | 3.61 ± 0.00 * | n.d. | 32.10 ± 0.01 | 25.12 ± 0.54 | n.d. | 17.06 ± 1.05 |
| GESWCR | 1.12 ± 0.02 * | 2.43 ± 0.01 * | n.d. | n.d | n.d. | n.d. | n.d |
| DGLGYY | 0.63 ± 0.04 * | 3.83 ± 0.19 * | 82.60 ± 0.58 | n.d. | n.d. | n.d. | 20.90 ± 1.31 |
| MFTGPY | 0.94 ± 0.01 * | 2.44 ± 0.02 * | n.d. | n.d. | 15.79 ± 0.55 | 214.30 ± 2.83 | 12.49 ± 2.01 |
| SKDAPY | 0.43 ± 0.04 * | 1.06 ± 0.13 * | n.d. | n.d. | n.d. | n.d. | n.d |
| SQLPGW | 0.53 ± 0.01 * | 2.95 ± 0.24 * | 50.50 ± 0.95 | n.d. | n.d. | n.d. | 11.63 ± 1.06 |
| YDLHGY | 0.64 ± 0.05 * | 3.59 ± 0.46 * | n.d. | 10.60 ±0.01 | n.d. | n.d. | n.d |
| GSYHDSK | 0.23 ± 0.02 * | 1.74 ± 0.07 * | n.d. | n.d | 19.57 ± 0.54 | n.d. | n.d |
| NGENDWR | 0.13 ± 0.01 | 0.82 ± 0.05 | n.d. | n.d | n.d. | n.d. | n.d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Arana, N.; Jiménez-Aliaga, K.; Intiquilla, A.; León, J.A.; Flores, E.; Zavaleta, A.I.; Izaguirre, V.; Solis-Calero, C.; Hernández-Ledesma, B. Protection against Oxidative Stress and Metabolic Alterations by Synthetic Peptides Derived from Erythrina edulis Seed Protein. Antioxidants 2022, 11, 2101. https://doi.org/10.3390/antiox11112101
Rodríguez-Arana N, Jiménez-Aliaga K, Intiquilla A, León JA, Flores E, Zavaleta AI, Izaguirre V, Solis-Calero C, Hernández-Ledesma B. Protection against Oxidative Stress and Metabolic Alterations by Synthetic Peptides Derived from Erythrina edulis Seed Protein. Antioxidants. 2022; 11(11):2101. https://doi.org/10.3390/antiox11112101
Chicago/Turabian StyleRodríguez-Arana, Nathaly, Karim Jiménez-Aliaga, Arturo Intiquilla, José A. León, Eduardo Flores, Amparo Iris Zavaleta, Víctor Izaguirre, Christian Solis-Calero, and Blanca Hernández-Ledesma. 2022. "Protection against Oxidative Stress and Metabolic Alterations by Synthetic Peptides Derived from Erythrina edulis Seed Protein" Antioxidants 11, no. 11: 2101. https://doi.org/10.3390/antiox11112101