Characterizing Industrial and Artisanal Fishing Vessel Catch Composition Using Environmental DNA and Satellite-Based Tracking Data
Abstract
:1. Introduction
2. Materials and Methods
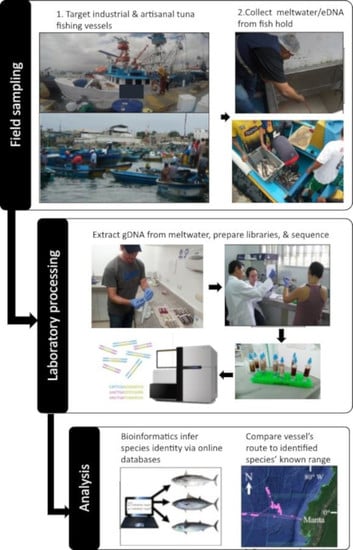
2.1. Field Sites and Vessel Sampling
2.2. eDNA Extraction and Library Preparation
2.3. Read Processing and Bioinformatics
2.4. Pairing Industrial Vessel Route and Metabarcoding Results
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture (2020) Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Agnew, D.J.; Pearce, J.; Pramod, G.; Peatman, T.; Watson, R.; Beddington, J.R.; Pitcher, T.J. Estimating the worldwide extent of illegal fishing. PLoS ONE 2009, 4, e4570. [Google Scholar] [CrossRef] [Green Version]
- Béné, C.; Barange, M.; Subasinghe, R.; Pinstrup-Andersen, P.; Merino, G.; Hemre, G.I.; Williams, M. Feeding 9 billion by 2050–Putting fish back on the menu. Food Secur. 2015, 7, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Al-Abdulrazzak, D.; Galland, G.R.; McClenachan, L.; Hocevar, J. Opportunities for improving global marine conservation through multilateral treaties. Mar. Policy 2017, 86, 247–252. [Google Scholar] [CrossRef]
- Willette, D.A.; Cheng, S.H. Delivering on seafood traceability under the new US import monitoring program. Ambio 2018, 47, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. Fisheries cooperation in the South China Sea: Evaluating the options. Mar. Policy 2018, 89, 67–76. [Google Scholar] [CrossRef]
- He, J. Do unilateral trade measures really catalyze multilateral environmental agreements? Int. Environ. Agreem. Politics Law Econ. 2019, 19, 577–593. [Google Scholar] [CrossRef]
- FAO. The State of the World Fisheries and Aquaculture 2016—Contributing to Food Security and Nutrition for All; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016. [Google Scholar]
- Ayala Villa, B. Pesca ilegal, no declara, no reglamentada—Subsecretaría de Recursos Pesqueros. In En el Seminario: Biotecnología Aplicada a la Lucha Contra la Pesca Ilegal, no Declarada y no Reglamentada en los Países del Océano Pacífico; ESPOL Polytechnic University: Guayaquil, Ecuador, 2017. [Google Scholar]
- Bucaram, S.J.; Hearn, A.; Trujillo, A.M.; Rentería, W.; Bustamante, R.H.; Morán, G.; Reck, G.; García, J.L. Assessing fishing effects inside and outside an MPA: The impact of the Galapagos Marine Reserve on the Industrial pelagic tuna fisheries during the first decade of operation. Mar. Policy 2018, 87, 212–225. [Google Scholar] [CrossRef]
- Alava, J.J.; Lindop, A.; Jacquet, J. Marine fisheries catch reconstructions for continental Ecuador: 1950–2010. In Fisheries Centre Working Paper; Fisheries Centre, University of British Columbia: Vancouver, BC, Canada, 2015; Volune 34, p. 25. [Google Scholar]
- Alava, J.J.; Tatar, B.; Barragán, M.J.; Castro, C.; Rosero, P.; Denkinger, J.; Jiménez, P.J.; Carvajal, R.; Samaniego, J. Mitigating cetacean bycatch in coastal Ecuador: Governance challenges for small-scale fisheries. Mar. Policy 2019, 110, 102769. [Google Scholar] [CrossRef]
- Schwartz, M.K.; Luikart, G.; Waples, R.S. Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 2007, 22, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourlat, S.J.; Borja, A.; Gilbert, J.; Taylor, M.I.; Davies, N.; Weisberg, S.B.; Griffith, J.F.; Lettieri, T.; Field, D.; Benzie, J.; et al. Genomics in marine monitoring: New opportunities for assessing marine health status. Mar. Pollut. Bull. 2013, 74, 19–31. [Google Scholar] [CrossRef]
- Lu, G.; Luo, M. Genomes of major fishes in world fisheries and aquaculture: Status, application and perspective. Aquac. Fish. 2020, 5, 163–173. [Google Scholar] [CrossRef]
- Marko, P.B.; Lee, S.C.; Rice, A.M.; Gramling, J.M.; Fitzhenry, T.M.; McAlister, J.S.; Moran, A.L. Mislabelling of a depleted reef fish. Nature 2004, 430, 309–310. [Google Scholar] [CrossRef]
- Clark, L.F. The current status of DNA barcoding technology for species identification in fish value chains. Food Policy 2015, 54, 85–94. [Google Scholar] [CrossRef]
- Luque, G.M.; Donlan, C.J. The characterization of seafood mislabeling: A global meta-analysis. Biol. Conserv. 2019, 236, 556–570. [Google Scholar] [CrossRef]
- Shehata, H.R.; Bourque, D.; Steinke, D.; Chen, S.; Hanner, R. Survey of mislabelling across finfish supply chain reveals mislabelling both outside and within Canada. Food Res. Int. 2019, 121, 723–729. [Google Scholar] [CrossRef]
- Barendse, J.; Roel, A.; Longo, C.; Andriessen, L.; Webster, L.M.; Ogden, R.; Neat, F. DNA barcoding validates species labelling of certified seafood. Curr. Biol. 2019, 29, R198–R199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willette, D.A.; Esteves, S.C.; Fitzpatrick, B.; Smith, M.L.; Wilson, K.; Yuan, X. The last mile challenge: Certified seafood and federal labeling laws out of sync at the end of the supply chain in Los Angeles, California. Mar. Policy 2021, 125, 104380. [Google Scholar] [CrossRef]
- Asis, A.M.J.M.; Lacsamana, J.K.M.; Santos, M.D. Illegal trade of regulated and protected aquatic species in the Philippines detected by DNA barcoding. Mitochondrial DNA Part A 2016, 27, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, C.A.; Potts, R.W.; Walsh, M.B.; Usher, J.; Griffiths, A.M. Using DNA barcoding to investigate patterns of species utilisation in UK shark products reveals threatened species on sale. Sci. Rep. 2019, 9, 1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardo, C.; de Lima Adachi, A.M.C.; da Cruz, V.P.; Foresti, F.; Loose, R.H.; Bornatowski, H. The label “Cação” is a shark or a ray and can be a threatened species! Elasmobranch trade in Southern Brazil unveiled by DNA barcoding. Mar. Policy 2020, 116, 103920. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Yu, D.W.; de Bruyn, M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.P.; Port, J.; Yamahara, K.M.; Martone, R.G.; Lowell, N.; Thomsen, P.F.; Mach, M.E.; Bennett, M.; Prahler, E.; Caldwell, M.R.; et al. Harnessing DNA to improve environmental management. Science 2014, 344, 1455–1456. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Fronhofer, E.A.; Mächler, E.; Altermatt, F. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Biorxiv 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.O.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Arakiet, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2014, 183, 4–18. [Google Scholar] [CrossRef]
- Minamoto, T.; Yamanaka, H.; Takahara, T.; Honjo, M.N.; Kawabata, Z. Surveillance of fish species composition using environmental DNA. Limnology 2011, 13, 193–197. [Google Scholar] [CrossRef] [Green Version]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z. Estimation of fish biomass using environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef] [Green Version]
- Pikitch, E.K. A tool for finding rare marine species. Science 2018, 360, 1180–1182. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H.; Hajibabaei, M.; Burns, J.M.; Hallwachs, W.; Remigio, E.; Hebert, P.D. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B Biol. 2005, 360, 1835–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardura, A.; Planes, S.; Garcia-Vazquez, E. Applications of DNA barcoding to fish landings: Authentication and diversity assessment. Zookeys 2013, 365, 49–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerde, C.L. Can we manage fisheries with the inherent uncertainty from eDNA? J. Fish Biol. 2021, 98, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Rourke, M.L.; Fowler, A.M.; Hughes, J.M.; Broadhurst, M.K.; DiBattista, J.D.; Fielder, S.; Furlan, E.M. Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 2021. [Google Scholar] [CrossRef]
- Stoeckle, M.Y.; Adolf, J.; Charlop-Powers, Z.; Dunton, K.J.; Hinks, G.; VanMorter, S.M. Trawl and eDNA assessment of marine fish diversity, seasonality, and relative abundance in coastal New Jersey, USA. ICES J. Mar. Sci. 2021, 78, 293–304. [Google Scholar] [CrossRef]
- Burgess, M.G.; McDermott, G.R.; Owashi, B.; Reeves, L.E.P.; Clavelle, T.; Ovando, D.; Costello, C. Protecting marine mammals, turtles, and birds by rebuilding global fisheries. Science 2018, 359, 1255–1258. [Google Scholar] [CrossRef] [Green Version]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Dillon, M.R.; Zhang, Y.; Rideout, J.R.; Bolyen, E.; Li, H.; Caporaso, J.G. q2-longitudinal: Longitudinal and paired-sample analyses of microbiome data. MSystems 2018, 3, e00219-18. [Google Scholar] [CrossRef] [Green Version]
- Aguirre-Pabon, J.C.; Orozco Berdugo, G., Jr.; Narváez Barandica, J.C. Genetic status, source and establishment risk of the giant tiger shrimp (Penaeidae: Penaeus monodon), an invasive species in Colombian Caribbean waters. Acta Biológica Colomb. 2015, 20, 117–127. [Google Scholar] [CrossRef]
- Spens, J.; Evans, A.R.; Halfmaerten, D.; Knudsen, S.W.; Sengupta, M.E.; Mak, S.S.; Hellström, M. Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: Advantage of enclosed filter. Methods Ecol. Evol. 2017, 8, 635–645. [Google Scholar] [CrossRef] [Green Version]
- Gold, Z.; Sprague, J.; Kushner, D.J.; Zerecero Marin, E.; Barber, P.H. eDNA metabarcoding as a biomonitoring tool for marine protected areas. PLoS ONE 2021, 16, e0238557. [Google Scholar] [CrossRef] [PubMed]
- Curd, E.E.; Gold, Z.; Kandlikar, G.S.; Gomer, J.; Ogden, M.; O’Connell, T.; Pipes, L.; Schweizer, T.M.; Rabichow, L.; Lin, M.; et al. Anacapa Toolkit: An environmental DNA toolkit for processing multilocus metabarcode datasets. Methods Ecol. Evol. 2019, 10, 1469–1475. [Google Scholar] [CrossRef] [Green Version]
- Kandlikar, G.S.; Gold, Z.J.; Cowen, M.C.; Meyer, R.S.; Freise, A.C.; Kraft, N.J.; Curd, E.E. ranacapa: An R package and Shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000Research 2018, 7, 1734. [Google Scholar] [CrossRef] [PubMed]
- McCauley, D.J.; Woods, P.; Sullivan, B.; Bergman, B.; Jablonicky, C.; Roan, A.; Hirshfield, M.; Boerder, K.; Worm, B. Ending hide and Seek at Sea. Science 2016, 351, 1148–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taconet, M.; Kroodsma, D.A.; Fernandes, J.A. Global Atlas of AIS-Based Fishing Activity—Challenges and Opportunities; FAO: Rome, Italy, 2019. [Google Scholar]
- Bez, N.; Walker, E.; Gaertner, D.; Rivoirard, J.; Gaspar, P. Fishing activity of tuna purse seiners estimated from vessel monitoring system (VMS) data. Can. J. Fish. Aquat. Sci. 2011, 68, 1998–2010. [Google Scholar] [CrossRef]
- de Souza, E.N.; Boerder, K.; Matwin, S.; Worm, B. Improving fishing pattern detection from satellite AIS using data mining and machine learning. PLoS ONE 2016, 11, e0158248. [Google Scholar]
- Kaschner, K.; Kesner-Reyes, K.; Garilao, C.; Segschneider, J.; Rius-Barile, J.; Rees, T.; Froese, R. AquaMaps: Predicted Range Maps for Aquatic Species. Available online: https://www.aquamaps.org (accessed on 20 February 2021).
- von Gersdorff Jørgensen, L.; Nielsen, J.W.; Villadsen, M.K.; Vismann, B.; Dalvin, S.; Mathiessen, H.; Buchmann, K. A non-lethal method for detection of Bonamia ostreae in flat oyster (Ostrea edulis) using environmental DNA. Sci. Rep. 2020, 10, 16143. [Google Scholar] [CrossRef]
- Noh, E.S.; Lee, M.N.; Kim, E.M.; Nam, B.H.; Noh, J.K.; Park, J.Y.; Kang, J.H. Discrimination of raw material species in mixed seafood products (surimi) using the next generation sequencing method. Food Biosci. 2021, 41, 100786. [Google Scholar] [CrossRef]
- Willette, D.A.; Simmonds, S.E.; Cheng, S.H.; Esteves, S.; Kane, T.L.; Nuetzel, H.; Pilaud, N.; Rachmawati, R.; Barber, P.H. Using DNA barcoding to track seafood mislabeling in Los Angeles restaurants. Conserv. Biol. 2017, 31, 1076–1085. [Google Scholar]
- Leray, M.; Boehm, J.T.; Mills, S.C.; Meyer, C.P. Moorea BIOCODE barcode library as a tool for understanding predator–prey interactions: Insights into the diet of common predatory coral reef fishes. Coral Reefs 2012, 31, 383–388. [Google Scholar] [CrossRef]
- Gold, Z.; Curd, E.; Goodwin, K.; Choi, E.; Frable, B.; Thompson, A.; Barber, P. Improving Metabarcoding Taxonomic Assignment: A Case Study of Fishes in a Large Marine Ecosystem. Authorea Prepr. 2021. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. (Eds.) FishBase; World Wide Web Electronic Publication; 2021; Available online: www.fishbase.org (accessed on 20 February 2021).
- Béné, C. Small-Scale Fisheries: Assessing Their Contribution to Rural Livelihoods in Developing Countries; FAO Fisheries Circular No. 1008; FAO: Rome, Italy, 2006; p. 46. [Google Scholar]
- Peckham, S.H.; Díaz, D.M.; Walli, A.; Ruiz, G.; Crowder, L.B.; Nichols, W.J. Small-scale fisheries bycatch jeopardizes endangered Pacific loggerhead turtles. PLoS ONE 2007, 2, e1041. [Google Scholar] [CrossRef] [PubMed]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol. Ecol. Resour. 2014, 14, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Moyer, G.R.; Diaz-Ferguson, E.; Hill, J.E.; Shea, C. Assessing environmental DNA detection in controlled lentic systems. PLoS ONE 2014, 9, e103767. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Bernatchez, L. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Di Muri, C.; Lawson Handley, L.; Bean, C.W.; Li, J.; Peirson, G.; Sellers, G.S.; Hänfling, B. Read counts from environmental DNA (eDNA) metabarcoding reflect fish abundance and biomass in drained ponds. Metabarcoding Metagenomics 2020, 4, 97–112. [Google Scholar] [CrossRef]
- Kelly, R.P.; Shelton, A.O.; Gallego, R. Understanding PCR processes to draw meaningful conclusions from environmental DNA studies. Sci. Rep. 2019, 9, 12133. [Google Scholar] [CrossRef] [Green Version]
- McLaren, M.R.; Willis, A.D.; Callahan, B.J. Consistent and correctable bias in metagenomic sequencing experiments. Elife 2019, 8, e46923. [Google Scholar] [CrossRef] [PubMed]
- Beng, K.C.; Corlett, R.T. Applications of environmental DNA (eDNA) in ecology and conservation: Opportunities, challenges and prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Bernatchez, L.; Wellenreuther, M.; Araneda, C.; Ashton, D.T.; Barth, J.M.; Beacham, T.D.; Withler, R.E. Harnessing the power of genomics to secure the future of seafood. Trends Ecol. Evol. 2017, 32, 665–680. [Google Scholar] [CrossRef] [Green Version]
- Sigsgaard, E.E.; Jensen, M.R.; Winkelmann, I.E.; Møller, P.R.; Hansen, M.M.; Thomsen, P.F. Population-level inferences from environmental DNA—Current status and future perspectives. Evol. Appl. 2020, 13, 245–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigsgaard, E.E.; Nielsen, I.B.; Bach, S.S.; Lorenzen, E.D.; Robinson, D.P.; Knudsen, S.W.; Thomsen, P.F. Population characteristics of a large whale shark aggregation inferred from seawater environmental DNA. Nat. Ecol. Evol. 2017, 1, 4. [Google Scholar] [CrossRef]
- Parsons, K.M.; Everett, M.; Dahlheim, M.; Park, L. Water, water everywhere: Environmental DNA can unlock population structure in elusive marine species. R. Soc. Open Sci. 2018, 5, 180537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinholdt Jensen, M.; Egelyng Sigsgaard, E.; Agersnap, S.; Jessen Rasmussen, J.; Baattrup-Pedersen, A.; Wiberg-Larsen, P.; Francis Thomsen, P. Seasonal turnover in community composition of stream-associated macroinvertebrates inferred from freshwater environmental DNA metabarcoding. Environ. DNA 2021, 1–16. [Google Scholar] [CrossRef]
- Pochardt, M.; Allen, J.M.; Hart, T.; Miller, S.D.; Yu, D.W.; Levi, T. Environmental DNA facilitates accurate, inexpensive, and multiyear population estimates of millions of anadromous fish. Mol. Ecol. Resour. 2020, 20, 457–467. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Côté, G.; Leclerc, V.; Bernatchez, L. Quantifying relative fish abundance with eDNA: A promising tool for fisheries management. J. Appl. Ecol. 2016, 53, 1148–1157. [Google Scholar] [CrossRef]
- Ames, C.L.; Ohdera, A.H.; Colston, S.M.; Collins, A.G.; Fitt, W.K.; Morandini, A.C.; Vora, G.J. Fieldable Environmental DNA Sequencing to Assess Jellyfish Biodiversity in Nearshore Waters of the Florida Keys, United States. Front. Mar. Sci. 2021, 8, 369. [Google Scholar] [CrossRef]


| Order | Family | Genus | Species (Common Name) |
|---|---|---|---|
| Acanthuriformes | Scianenidae | - | - |
| Beloniformes | Exocoetidae | Exocoetus | Exocoetus volitans (two-wing flying fish) |
| ““ | Hemiramphidae | Oxyporhamphus | Oxyporhamphus Micropterus (bigwing halfbeak) |
| Carangiformes | Carangidae | Seriola | Seriola rivoliana (longfin yellowtail) |
| ““ | ““ | Chloroscombrus | Chloroscombrus orquesta (Pacific bumper) |
| ““ | ““ | Decapterus | Decapterus macrosoma (shortfin scad) |
| Gadiformes | Merlucciidae | Merluccius | Merluccius productus (North Pacific hake) |
| Istiophoriformes | Istiophridae | - | - |
| Scombriformes | Scombridae | Acanthocybium | Acanthocybium solandri (wahoo) |
| ““ | ““ | Auxis | Auxis rochei (bullet tuna) |
| ““ | ““ | ““ | Auxis thazard (frigate tuna) |
| ““ | ““ | Katsuwonus | Katsuwonus pelamis (skipjack tuna) |
| ““ | ““ | Sarda | Sarda orientalis (striped bonito) |
| ““ | ““ | Euthynnus | - |
| ““ | ““ | Thunnus | - |
| Tetradontiformes | Balistidae | Canthidermis | Canthidermis maculate (rough triggerfish) |
| Artisanal Fishing Vessel | Industrial Fishing Vessel | |||||
|---|---|---|---|---|---|---|
| Species | A_AL | A_EC | A_ED | I_DA | I_EC | I_LZ |
| Acanthocybium solandri | P | |||||
| Auxis rochei | P | P | ||||
| Auxis thazard | P | P | P | |||
| Canthidermis maculate | P | |||||
| Chloroscombrus sorqueta | P | |||||
| Decapterus macrosoma | P | |||||
| Exocoetus volitans | P | |||||
| Katsuwonus pelamis | P | P | P | |||
| Merluccius productus | P | |||||
| Oxyporhamphus Micropterus | P | |||||
| Sarda orientalis | P | |||||
| Seriola rivoliana | P | |||||
| Genus level only | ||||||
| Euthynnus | P | |||||
| Thunnus | P | P | P | P | ||
| Species | A_AL | A_EC | A_ED | I_DA | I_EC | I_LZ |
|---|---|---|---|---|---|---|
| Acanthocybium solandri (peje sierra) | P | P | ||||
| Coryphaena hippurus (dorado) | P | P | ||||
| Euthynnus lineatus (pata seca) | P | |||||
| Istiompax indica (picudo) | P | |||||
| Katsuwonus pelamis (bonito) | P | P | P | P | ||
| Merluccius gayi (merluza) | P | |||||
| Thunnus alalunga (albacora) | P | P | P | P | P | P |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willette, D.A.; Navarrete-Forero, G.; Gold, Z.; Lizano, A.M.D.; Gonzalez-Smith, L.; Sotil, G. Characterizing Industrial and Artisanal Fishing Vessel Catch Composition Using Environmental DNA and Satellite-Based Tracking Data. Foods 2021, 10, 1425. https://doi.org/10.3390/foods10061425
Willette DA, Navarrete-Forero G, Gold Z, Lizano AMD, Gonzalez-Smith L, Sotil G. Characterizing Industrial and Artisanal Fishing Vessel Catch Composition Using Environmental DNA and Satellite-Based Tracking Data. Foods. 2021; 10(6):1425. https://doi.org/10.3390/foods10061425
Chicago/Turabian StyleWillette, Demian A., Gabriela Navarrete-Forero, Zachary Gold, Apollo Marco D. Lizano, Leonardo Gonzalez-Smith, and Giovanna Sotil. 2021. "Characterizing Industrial and Artisanal Fishing Vessel Catch Composition Using Environmental DNA and Satellite-Based Tracking Data" Foods 10, no. 6: 1425. https://doi.org/10.3390/foods10061425