Gradual Distance Dispersal Shapes the Genetic Structure in an Alpine Grasshopper
Abstract
:1. Introduction
2. Material and Methods
2.1. Species and Sampling
2.2. DNA Extraction and Sequencing
2.3. Estimation of Genetic Diversity
2.4. Migration Model Selection and Estimation of Population Genetics Parameters
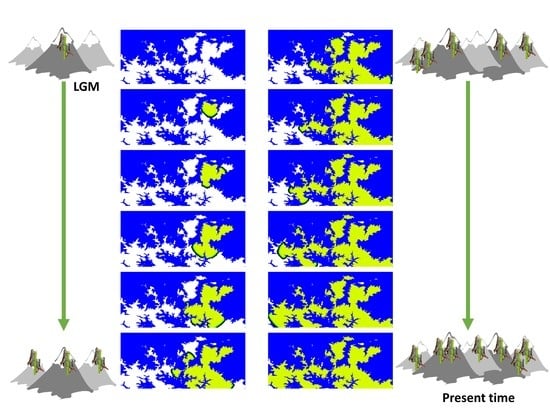
2.4.1. Spatially-Explicit Computer Simulations
2.4.2. Summary Statistics
2.4.3. Selection among Migration Models
2.4.4. Estimation of Population Genetic and Environmental Parameters
3. Results
3.1. Genetic Diversity
3.2. Selection of the Best-Fitting Migration Model
3.3. Estimation of Population Genetics and Environmental Parameters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Archiving
References
- Hewitt, G.M. Post-glacial recolonization of European biota. Biol. J. Linn. Soc. 1999, 68, 87–112. [Google Scholar] [CrossRef]
- Hewitt, G.M. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Knowles, L.L. Did the Pleistocene glaciations promote divergence? Tests of explicit refugial models in montane grasshoppers. Mol. Ecol. 2001, 10, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Greca, L. Le Cophopodisma (Ort. Catantopidae) dell’Appennino ed il loro differenziamento infraspecifico. Ann. Istit. Museo Zool. Univ. Napoli. 1954, 6, 1–20. [Google Scholar]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.; Cosson, J. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B. 2004, 359, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Weiss, S.; Ferrand, N. Phylogeography of Southern European Refugia: Evolutionary Perspectives on the Origins and Conservation of European Biodiversity; Springer: Berlin, Germany, 2007. [Google Scholar]
- Listl, D.; Poschlod, P.; Reisch, C. Phylogeography of a tough rock survivor in European dry grasslands. Plos One 2017, 12, e0179961. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, T. Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front. Zool. 2007, 4, 11. [Google Scholar] [CrossRef]
- Noguerales, V.; Cordero, P.J.; Ortego, J. Hierarchical genetic structure shaped by topography in a narrow-endemic montane grasshopper. Bmc Evol. Biol. 2016, 16, 96. [Google Scholar] [CrossRef]
- Noguerales, V.; García-Navas, V.; Cordero, P.J.; Ortego, J. The role of environment and core-margin effects on range-wide phenotypic variation in a montane grasshopper. J. Evol. Biol. 2016, 29, 2129–2142. [Google Scholar] [CrossRef] [Green Version]
- Kolář, F.; Dušková, E.; Sklenář, P. Niche shifts and range expansions along cordilleras drove diversification in a high-elevation endemic plant genus in the tropical Andes. Mol. Ecol. 2016, 25, 4593–4610. [Google Scholar] [CrossRef] [PubMed]
- Price, T.D.; Hooper, D.M.; Buchanan, C.D.; Johansson, U.S.; Tietze, D.T.; Alström, P.; Olsson, U.; Ghosh-Harihar, M.; Ishtiaq, F.; Gupta, S.K.; et al. Niche filling slows the diversification of Himalayan songbirds. Nature 2014, 509, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Quintero, I.; Jetz, W. Global elevational diversity and diversification of birds. Nature 2018, 555, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Sømme, L. Adaptations of terrestrial arthropods to the alpine environment. Biol. Rev. 1989, 64, 367–407. [Google Scholar] [CrossRef]
- Laiolo, P.; Obeso, J.R. Life-History Responses to the Altitudinal Gradient. In High Mountain Conservation in a Changing World. Advances in Global Change Research; Catalan, J., Ninot, J.M., Aniz, M., Eds.; Springer: Cham, Germany, 2017; Volume 25, pp. 127–173. [Google Scholar]
- Roff, D.A. The evolution of flightlessness in insects. Ecol. Monograph. 1990, 60, 389–421. [Google Scholar] [CrossRef]
- Wagner, D.L.; Liebherr, J.K. Flightlessness in insects. Trends Ecol. Evol. 1992, 7, 216–220. [Google Scholar] [CrossRef]
- Zera, A.J.; Harshman, L.G. The physiology of life history trade-offs in animals. Annu. Rev. Ecol. Syst 2001, 32, 95–126. [Google Scholar] [CrossRef]
- Trewick, S.A.; Wallis, G.P.; Morgan-Richards, M. Phylogeographical pattern correlates with Pliocene mountain building in the alpine scree weta (Orthoptera, Anostostomatidae). Mol. Ecol. 2000, 9, 657–666. [Google Scholar] [CrossRef]
- Levins, R. Evolution in changing environments. Some threoretical explorations. Monographs in Population Biology-2; Princeton University Press: Princeton, NJ, USA, 1968. [Google Scholar]
- Dussex, N.; Chuah, A.; Waters, J.M. Genome-wide SNPs reveal fine-scale differentiation among wingless alpine stonefly populations and introgression between winged and wingless forms. Evolution 2016, 70, 38–47. [Google Scholar] [CrossRef]
- Arenas, M.; Ray, N.; Currat, M.; Excoffier, L. Consequences of Range Contractions and Range Shifts on Molecular Diversity. Mol. Biol. Evol. 2012, 29, 207–218. [Google Scholar] [CrossRef]
- Knowles, L.L.; Massatti, R. Distributional shifts – not geographic isolation – as a probable driver of montane species divergence. Ecography 2017, 40, 1475–1485. [Google Scholar] [CrossRef]
- Alves, I.; Arenas, M.; Currat, M.; Sramkova Hanulova, A.; Sousa, V.C.; Ray, N.; Excoffier, L. Long-Distance Dispersal Shaped Patterns of Human Genetic Diversity in Eurasia. Mol. Biol. Evol. 2016, 33, 946–958. [Google Scholar] [CrossRef]
- Flø, D.; Ha, S. Aerial dispersal of invertebrates and mosses close to a receding alpine glacier in southern Norway. Arct. Antarct. Alp. Res. 2013, 45, 481–490. [Google Scholar] [CrossRef]
- Wada, S.; Kawakami, K.; Chiba, S. Snails can survive passage through a bird’s digestive system. J. Biogeogr. 2011, 39, 69–73. [Google Scholar] [CrossRef]
- Nogales, M.; López, H.; Emerson, B.C. How can large flightless beetles disperse by flight? The role of the omnivorous gulls on an oceanic island. In Proceedings of the Island Biology 2014, Honolulu, HI, USA, 7–11 July 2014. [Google Scholar]
- Jordano, P. What is long-distance dispersal? And a taxonomy of dispersal events. J. Ecol. 2017, 105, 75–84. [Google Scholar] [CrossRef]
- Thuiller, W.; Lavorel, S.; Araújo, M.B.; Sykes, M.T.; Prentice, I.C. Climate change threats to plant diversity in Europe. Proc. Natl. Acad. Sci. USA 2005, 102, 8245–8250. [Google Scholar] [CrossRef] [Green Version]
- Lawler, J.J.; Shafe, S.L.; White, D.; Kareiva, P.; Maurer, E.P.; Blaustein, A.R.; Bartlein, P.J. Projected climate-induced faunal change in the Western Hemisphere. Ecology 2009, 90, 588–597. [Google Scholar] [CrossRef]
- Zink, R.M.; Barrowclough, G.F. Mitochondrial DNA under siege in avian phylogeography. Mol. Ecol. 2008, 17, 2107–2121. [Google Scholar] [CrossRef]
- García-Olivares, V.; López, H.; Patiño, J.; Alvarez, N.; Machado, A.; Carracedo, J.C.; Soler, V.; Emerson, B.C. Evidence for mega-landslides as drivers of island colonization. J. Biogeogr. 2017, 44, 1053–1064. [Google Scholar] [CrossRef] [Green Version]
- Laiolo, P.; Obeso, J.R. Plastic responses to temperature vs. local adaptation at the cold extreme of the climate gradient. Evol. Biol. 2015, 42, 473–482. [Google Scholar] [CrossRef]
- Laiolo, P.; Pato, J.; Obeso, J.R. Ecological and evolutionary drivers of the elevational gradient of diversity. Ecol. Lett. 2018, 21, 1022–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laiolo, P.; Illera, J.C.; Obeso, J.R. Local climate determines intra-and interspecific variation in sexual size dimorphism in mountain grasshopper communities. J. Evol. Biol. 2013, 26, 2171–2183. [Google Scholar] [CrossRef] [PubMed]
- Laiolo, P.; Illera, J.C.; Meléndez, L.; Segura, A.; Obeso, J.R. Abiotic, biotic and evolutionary control of the distribution of C and N isotopes in food webs. Am. Nat. 2015, 185, 169–182. [Google Scholar] [CrossRef]
- Folmer, O.; Blac, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- Pato, J.; Illera, J.C.; Obeso, J.R.; Laiolo, P. The roles of geography, climate and sexual selection in driving divergence among insect populations in mountaintops. J. Biogeogr. 2019, 46, 784–795. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nei, M. Molecular evolutionary genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Tajima, F. The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 1996, 143, 1457–1465. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Currat, M.; Arenas, M.; Quilodràn, C.S.; Excoffier, K.; Ray, N. SPLATCHE3: Simulation of serial genetic data under spatially explicit evolutionary scenarios including long-distance dispersal. Bioinformatics 2019, btz311. [Google Scholar] [CrossRef]
- Ray, N.; Excoffier, L. A first step towards inferring levels of long-distance dispersal during past expansions. Mol. Ecol. Res. 2010, 10, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Lao, D.J.; Ruiz-Zapata, M.B.; Gil-García, M.J.; Ballesteros, D.; Jiménez-Sanchez, M. Palaeoenvironmental research at Rexidora Cave: New evidence of cold and dry conditions in NW Iberia during MIS 3. Quatern. Int. 2015, 379, 35–46. [Google Scholar] [CrossRef]
- Serrano, E.; González-Trueba, J.J.; Pellitero, R.; Gómez-Lende, M. Quaternary glacial history of the Cantabrian Mountains of northern Spain: A new synthesis. In Quaternary Glaciation in the Mediterranean Mountains; Hughes, P.D., Woodward, J.C., Eds.; Geological Society: London, UK, 2017; Volume 433, pp. 55–85. [Google Scholar]
- Novembre, J.; Galvani, A.P.; Slatkin, M. The geographic spread of the CCR5 Delta32 HIV-resistance allele. Plos Biol. 2005, 3, e339. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, L.H.; Strazanac, J.S.; Roderick, G.K. Molecular phylogeny of Banza (Orthoptera: Tettigoniidae), the endemic katydids of the Hawaiian Archipelago. Mol. Phyl. Evol. 2006, 41, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, D.; Leuenberger, C.; Excoffier, L. Efficient approximate Bayesian computation coupled with Markov Chain Monte Carlo without likelihood. Genetics 2009, 182, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, D.; Leuenberger, C.; Neuenschwander, S.; Excoffier, L. ABCtoolbox: A versatile toolkit for approximate Bayesian computations. Bmc Bioinform. 2010, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A. Approximate Bayesian Computation in Evolution and Ecology. Annu. Rev. Ecol. Evol. Syst 2010, 41, 379–406. [Google Scholar] [CrossRef]
- Bertorelle, G.; Benazzo, A.; Mona, S. ABC as a flexible framework to estimate demography over space and time: Some cons, many pros. Mol. Ecol. 2010, 19, 2609–2625. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Seielstad, M.T.; Perez-Lezaun, A.; Feldman, M.W. Population growth of human Y chromosomes: A study of Y chromosome microsatellites. Mol. Biol. Evol. 1999, 16, 1791–1798. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Csillery, K.; François, O.; Blum, M.G. abc: An R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 2012, 3, 475–479. [Google Scholar] [CrossRef]
- Illera, J.C.; Palmero, A.M.; Laiolo, P.; Rodríguez, F.; Moreno, Á.C.; Navascués, M. Genetic, morphological, and acoustic evidence reveals lack of diversification in the colonization process in an island bird. Evolution 2014, 68, 2259–2274. [Google Scholar] [CrossRef]
- Padilla, D.P.; Spurgin, L.G.; Fairfield, E.A.; Illera, J.C.; Richardson, D.S. Population history, gene flow and bottlenecks in island populations of a secondary seed disperser, the southern grey shrike (Lanius meridionalis koenigi). Ecol. Evol. 2015, 5, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Jalut, G.; Esteban Amat, A.; Bonnet, L.; Gauquelin, T.; Fontugne, M. Holocene climatic changes in the Western Mediterranean, from south-east France to south-east Spain. Palaeogeo. Palaeoclim. Palaeoecol. 2000, 160, 255–290. [Google Scholar] [CrossRef]
- Carrión, Y.; Kaal, J.; López-Sáez, J.A.; López-Merino, L.; Martínez Cortizas, A. Holocene vegetation changes in NW Iberia revealed by anthracological and palynological records from a colluvial soil. Holocene 2010, 20, 53–66. [Google Scholar] [CrossRef]
- Knowles, L.L.; Alvarado-Serrano, D.F. Exploring the population genetic consequences of the colonization process with spatio-temporally explicit models: Insights from coupled ecological, demographic and genetic models in montane grasshoppers. Mol. Ecol. 2010, 19, 3727–3745. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Knowles, L.L. Species-specific responses to island connectivity cycles: Refined models for testing phylogeographic concordance across a Mediterranean Pleistocene Aggregate Island Complex. Mol. Ecol. 2015, 24, 4252–4268. [Google Scholar] [CrossRef]
- García-Verdugo, C.; Caujapé-Castells, J.; Illera, J.C.; Mairal, M.; Patiño, J.; Reyes-Betancort, A.; Scholz, S. Pleistocene extinctions as drivers of biogeographical patterns on the easternmost Canary Islands. J. Biogeogr. 2019, 46, 845–859. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Knowles, L.L. Genomic tests of the species-pump hypothesis: Recent island connectivity cycles drive population divergence but not speciation in Caribbean crickets across the Virgin Islands. Evolution 2015, 69, 1501–1517. [Google Scholar] [CrossRef]
- Weigelt, P.; Steinbauer, M.J.; Sarmento Cabral, J.; Kreft, H. Late Quaternary climate change shapes island biodiversity. Nature 2016, 532, 99–102. [Google Scholar] [CrossRef]
- Knowles, L.L.; Richards, C.L. Importance of genetic drift during Pleistocene divergence as revealed by analyses of genomic variation. Mol. Ecol. 2005, 14, 4023–4032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewitt, G.M.; Nichols, R.A.; Barton, N.H. Homogamy in a hybrid zone in the alpine grasshopper Podisma pedestris. Heredity 1987, 59, 457–466. [Google Scholar] [CrossRef]
- Trakhtenbrot, A.; Nathan, R.; Perry, G.; Richardson, D.M. The importance of long-distance dispersal in biodiversity conservation. Divers. Distrib. 2005, 11, 173–181. [Google Scholar] [CrossRef]
- Mona, S.; Ray, N.; Arenas, M.; Excoffier, L. Genetic consequences of habitat fragmentation during a range expansion. Heredity 2014, 112, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, J.A.; Hong-Chang, L.; Dodd, J.L.; Williams, S.E. Comparison of Grasshopper (Orthoptera: Acrididae) Ecology on the Grasslands of the Asian Steppe in Inner Mongolia and the Great Plains of North America. J. Orthoptera Res. 1994, 2, 4–14. [Google Scholar] [CrossRef]
- Madrigal, L.; Posthumously, L.C.; Melendez-Obando, M.; Villegas-Palma, R.; Barrantes, R.; Raventos, H.; Pereira, R.; Luiselli, D.; Pettener, D.; Barbujani, G. High mitochondrial mutation rates estimated from deep-rooting Costa Rican pedigrees. Am. J. Phys. Anthr. 2012, 148, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Branco, C.; Velasco, M.; Benguigui, M.; Currat, M.; Ray, N.; Arenas, M. Consequences of diverse evolutionary processes on American genetic gradients of modern humans. Heredity 2018, 121, 548–556. [Google Scholar] [CrossRef]


| Locality | Acronym | Sample size | Group | Massif |
|---|---|---|---|---|
| Cantu l’Osu | COsu | 20 | 1 | Central |
| Campigüeños | Cam | 11 | 2 | Western |
| Llambria | Lla | 11 | 2 | Western |
| Tiatordos | Tia | 16 | 2 | Western |
| Maciédome | Mac | 18 | 2 | Western |
| Peña Ten | Ten | 15 | 3 | Western |
| Pileñes | Pil | 10 | 3 | Western |
| Cantu Cabroneru | CC | 15 | 4 | Central |
| Traviesos | Tra | 15 | 5 | Central |
| Cotalba | Cot | 15 | 5 | Central |
| Vega Ario | Va | 15 | 5 | Central |
| Vega Huerta | Vh | 16 | 5 | Central |
| Vegarredonda | VR | 17 | 5 | Central |
| Tiros Navarros | NV | 25 | 6 | Eastern |
| Peña Castil | PC | 16 | 6 | Eastern |
| Liordes | Lio | 6 | 6 | Eastern |
| Urriellu | U | 19 | 6 | Eastern |
| Camburero | Camb | 3 | 6 | Eastern |
| Morra Lechugales | MoHie | 19 | 7 | Eastern |
| Andara | A | 15 | 7 | Eastern |
| Casetón Andara | Ba | 15 | 7 | Eastern |
| Rasa | Ras | 15 | 7 | Eastern |
| Model | GDD | LDD | ||||||
|---|---|---|---|---|---|---|---|---|
| ABC approach | Pr | Rrej | Rreg | Rnn | Pr | Rrej | Rreg | Rnn |
| Probability | 0.97 | 0.87 | 1.00 | 1.00 | 0.03 | 0.13 | 0.00 | 0.00 |
| Parameter | ABC Approach | Mode | Mean | Median | 90% HPDI 1 |
|---|---|---|---|---|---|
| Time of the onset of the expansion 2 | Rejection | 19,336 | 17,547 | 17,558 | 15,256–19,775 |
| Ancestral population size | Neuralnet | 662 | 638 | 649 | 540–706 |
| Population growth rate | Regression | 0.28 | 0.28 | 0.28 | 0.27–0.29 |
| Migration rate | Rejection | 0.07 | 0.14 | 0.12 | 0.06–0.27 |
| Carrying capacity | Rejection | 122 | 177 | 154 | 105–313 |
| Mutation rate | Rejection | 1.37 × 10−6 | 4.78 × 10−6 | 4.66 × 10−6 | 5.15 ×10−7–9.43 × 10−6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Illera, J.C.; Arenas, M.; López-Sánchez, C.A.; Obeso, J.R.; Laiolo, P. Gradual Distance Dispersal Shapes the Genetic Structure in an Alpine Grasshopper. Genes 2019, 10, 590. https://doi.org/10.3390/genes10080590
Illera JC, Arenas M, López-Sánchez CA, Obeso JR, Laiolo P. Gradual Distance Dispersal Shapes the Genetic Structure in an Alpine Grasshopper. Genes. 2019; 10(8):590. https://doi.org/10.3390/genes10080590
Chicago/Turabian StyleIllera, Juan Carlos, Miguel Arenas, Carlos A. López-Sánchez, José Ramón Obeso, and Paola Laiolo. 2019. "Gradual Distance Dispersal Shapes the Genetic Structure in an Alpine Grasshopper" Genes 10, no. 8: 590. https://doi.org/10.3390/genes10080590