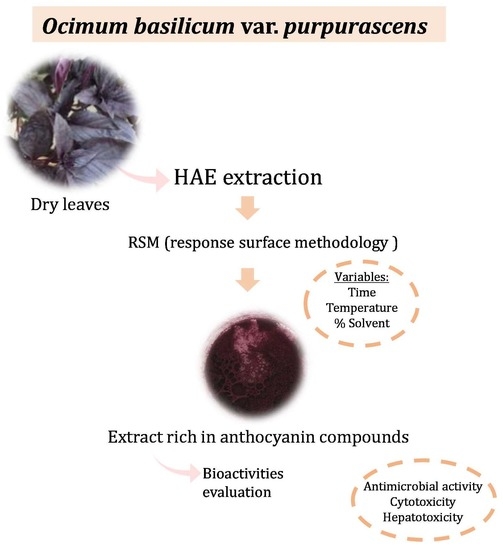
Optimization of the Extraction Process to Obtain a Colorant Ingredient from Leaves of Ocimum basilicum var. purpurascens
Abstract
:1. Introduction
2. Results
2.1. Response Criteria for the RSM Analysis
2.2. Theoretical Response Surface Models
2.3. Final Effects of the Studied Conditions of HAE on the Target Responses and Optimal Values that Maximize the Responses
2.4. Clustering of Anthocyanin Compounds According to the HAE Conditions that Maximize their Extraction
2.5. Dose-Response Analysis of the Solid-to-Liquid Effect at the Optimum Conditions
2.6. Evaluation of the Colorant Potential of the Extract Rich in Anthocyanin Compounds Obtained under Optimum Conditions from Leaves of O. basilicum var. purpurascens
2.7. Evaluation of the Bioactive Properties of the Extract Rich in Anthocyanin Compounds Obtained under Optimal Conditions from Leaves of O. basilicum var. purpurascens
2.7.1. Antimicrobial Activity
2.7.2. Cytotoxic Activity
3. Materials and Methods
3.1. Samples
3.2. Heat-Assisted Extraction
3.3. Calculation of the Extraction Yield
3.4. Chromatographic Analysis of Anthocyanin Compounds
3.5. Experimental Design, Modelling and Optimization
3.5.1. Experimental Design
3.5.2. Mathematical Modelling
3.5.3. Maximization of the Responses
3.6. Gropping the Responses by Cluster Analyses
3.7. Fitting Procedures and Statistical Analysis
3.8. Preparation of the Extract Rich in Anthocyanin Compounds Obtained under Optimum Conditions from the Leaves of O. basilicum var. purpurascens
3.9. Evaluation of the Colorant Potential of the Extract Rich in Anthocyanin Compounds Obtained under Optimum Conditions from the Leaves of O. basilicum var. purpurascens
3.10. Evaluation of the Bioactive Properties of the Extract Rich in Anthocyanin Compounds Obtained under Optimal Conditions from the Leaves of O. basilicum var. purpurascens.
3.10.1. Antimicrobial Activity
3.10.2. Cytotoxic Activity
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hoefkens, C.; Verbeke, W. Consumers’ health-related motive orientations and reactions to claims about dietary calcium. Nutrients 2013, 5, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Martins, N.; Roriz, C.L.; Morales, P.; Barros, L.; Ferreira, I.C.F.R. Food colorants: Challenges, opportunities and current desires of agro-industries to ensure consumer expectations and regulatory practices. Trends Food Sci. Technol. 2016, 52, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Amaya, D.B. Natural food pigments and colorants. Curr. Opin. Food Sci. 2016, 7, 20–26. [Google Scholar] [CrossRef]
- Neri-Numa, I.A.; Pessoa, M.G.; Paulino, B.N.; Pastore, G.M. Genipin: A natural blue pigment for food and health purposes. Trends Food Sci. Technol. 2017, 67, 271–279. [Google Scholar] [CrossRef]
- Almeida, H.H.S.; Barros, L.; Barreira, J.C.M.; Calhelha, R.C.; Heleno, S.A.; Sayer, C.; Miranda, C.G.; Leimann, F.V.; Barreiro, M.F.; Ferreira, I.C.F.R. Bioactive evaluation and application of different formulations of the natural colorant curcumin (E100) in a hydrophilic matrix (yogurt). Food Chem. 2018, 261, 224–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigurdson, G.T.; Tang, P.; Giusti, M.M. Natural Colorants: Food Colorants from Natural Sources. Annu. Rev. Food Sci. Technol 2017, 8, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Ananga, A.; Georgiev, V.; Ochieng, J.; Phills, B.; Tsolov, V. Production of Anthocyanins in Grape Cell Cultures: A Potential Source of Raw Material for Pharmaceutical, Food, and Cosmetic Industries. In The Mediterranean Genetic Code—Grapevine and Olive; IntechOpen: London, UK, 2013; pp. 247–287. [Google Scholar]
- Gerardi, C.; Tommasi, N.; Albano, C.; Blando, F.; Rescio, L.; Pinthus, E.; Mita, G. Prunus mahaleb L. fruit extracts: a novel source for natural food pigments. Eur Food Res. Technol. 2015, 241, 683–695. [Google Scholar] [CrossRef]
- Jornal Oficial da União Europeia. Regulamento (UE) N°1129/2011. Eur. Food Res. Technol. 2011, 25. [Google Scholar]
- Flanigan, P.M.; Niemeyer, E.D. Effect of cultivar on phenolic levels, anthocyanin composition, and antioxidant properties in purple basil (Ocimum basilicum L.). Food Chem. 2014, 164, 518–526. [Google Scholar] [CrossRef]
- Da Silva, F.J.; Nascimento, A.B.; Barbosa, L.N.; Magalhães, H.M. In vitro cultivation of purple basil Ocimum basilicum L. ‘red rubin’ at different levels of salts, charcoal, sucrose and potassium iodine. Aust. J. Crop Sci. 2017, 11, 1137–1145. [Google Scholar] [CrossRef]
- El-Ziat, R.A.; Swaefy, H.M.; Esmail, S.E.A. The Response of Red Rubin Basil Plant to Organic Fertilizer and Humic Acid versus Chemical Fertilizers. Middle East. J. Agric. Res. 2018, 7, 740–751. [Google Scholar]
- Alexandre, E.M.C.; Araújo, P.; Duarte, M.F.; de Freitas, V.; Pintado, M.; Saraiva, J.A. High-pressure assisted extraction of bioactive compounds from industrial fermented fig by-product. Int. J. Food Sci. Technol. 2017, 54, 2519–2531. [Google Scholar] [CrossRef]
- Alexandre, E.M.C.; Araújo, P.; Duarte, M.F.; de Freitas, V.; Pintado, M.; Saraiva, J.A. Experimental design, modeling, and optimization of high-pressure-assisted extraction of bioactive compounds from pomegranate peel. Food Bioprocess Tech. 2017, 10, 886–900. [Google Scholar] [CrossRef]
- Luna, M.C.; Bekhradi, F.; Ferreres, F.; Jordán, M.J.; Delshad, M.; Gil, M.I. Effect of Water Stress and Storage Time on Anthocyanins and Other Phenolics of Different Genotypes of Fresh Sweet Basil. J. Agric. Food Chem 2015, 63, 9223–9231. [Google Scholar] [CrossRef] [PubMed]
- Phippen, W.B.; Simon, J.E. Anthocyanins in Basil (Ocimum basilicum L.). J. Agric. Food Chem. 1998, 46, 1734–1738. [Google Scholar] [CrossRef]
- Ranic, M.; Nikolic, M.; Pavlovic, M.; Buntic, A.; Siler-Marinkovic, S.; Dimitrijevic-Brankovic, S. Optimization of microwave-assisted extraction of natural antioxidants from spent espresso coffee grounds by response surface methodology. J. Clean. Prod. 2014, 80, 69–79. [Google Scholar] [CrossRef]
- Sun, X.-H.; Zhou, T.-T.; Wei, C.-H.; Lan, W.-Q.; Zhao, Y.; Pan, Y.-J.; Wu, V.C.H. Antibacterial effect and mechanism of anthocyanin rich Chinese wild blueberry extract on various foodborne pathogens. Food Control 2018, 94, 155–161. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, H.; Yi, J.; Yang, B.; Li, M.; He, D.; Yang, W.; Zhang, Y.; Ni, H. Anti-tumor properties of anthocyanins from Lonicera caerulea ‘Beilei’ fruit on human hepatocellular carcinoma: In vitro and in vivo study. Biomed. Pharmacother. 2018, 104, 520–529. [Google Scholar] [CrossRef]
- Roriz, C.L.; Barros, L.; Prieto, M.A.; Morales, P.; Ferreira, I.C.F.R. Floral parts of Gomphrena globosa L. as a novel alternative source of betacyanins: Optimization of the extraction using response surface methodology. Food Chem. 2017, 229, 223–234. [Google Scholar] [CrossRef]
- Gonçalves, G.A.; Soares, A.A.; Correa, R.C.G.; Barros, L.; Haminiuk, C.W.I.; Peralta, R.M.; Ferreira, I.C.F.R.; Bracht, A. Merlot grape pomace hydroalcoholic extract improves the oxidative and inflammatory states of rats with adjuvant-induced arthritis. J. Funct. Foods 2017, 33, 408–418. [Google Scholar] [CrossRef] [Green Version]
- Vieira, V.; Prieto, M.A.; Barros, L.; Coutinho, J.A.P.; Ferreira, O.; Ferreira, I.C.F.R. Optimization and comparison of maceration and microwave extraction systems for the production of phenolic compounds from Juglans regia L. for the valorization of walnut leaves. Ind. Crops Prod. 2017, 107, 341–352. [Google Scholar] [CrossRef]
- Prieto, M.A.; Vázquez, J.A. In vitro determination of the lipophilic and hydrophilic antioxidant capacity of unroasted coffee bean extracts and their synergistic and antagonistic effects. Food Res. Int. 2014, 62, 1183–1196. [Google Scholar] [CrossRef] [Green Version]
- Pereira, E.; Antonio, A.L.; Barreira, J.C.M.; Barros, L.; Bento, A.; Ferreira, I.C.F.R. Gamma irradiation as a practical alternative to preserve the chemical and bioactive wholesomeness of widely used aromatic plants. Food Res. Int. 2015, 67, 338–348. [Google Scholar] [CrossRef] [Green Version]
- Carocho, M.; Barros, L.; Calhelha, R.C.; Ćirić, A.; Soković, M.; Santos-Buelga, C.; Morales, P.; Ferreira, I.C.F.R. Melissa officinalis L. decoctions as functional beverages: a bioactive approach and chemical characterization. Food Funct. 2015, 6, 2240–2248. [Google Scholar] [CrossRef] [PubMed]
- Barros, L.; Pereira, E.; Calhelha, R.C.; Dueñas, M.; Carvalho, A.M.; Santos-Buelga, C.; Ferreira, I.C.F.R. Bioactivity and chemical characterization in hydrophilic and lipophilic compounds of Chenopodium ambrosioides L. J. Funct. Foods 2013, 5, 1732–1740. [Google Scholar] [CrossRef]
Sample Availability: Samples are available from the authors. |





| Peak | Rt (min) | λmax (nm) | [M + H]+ | Main Fragment ESI- MSn [Intensity (%)] | Tentative Identification |
|---|---|---|---|---|---|
| P1 | 34.1 | 520 | 919 | 757(49),449(6),287(13) | Cyanidin-3-(p-coumaroyl-6′-caffeoyl)sophoroside isomer 1 A |
| P2 | 36.4 | 520 | 919 | 757(49),449(6),287(13) | Cyanidin-3-(p-coumaroyl-6′-caffeoyl)sophoroside isomer 2 A |
| P3 | 38.4 | 522 | 1005 | 757(6),535(11),287(11) | Cyanidin-3-(6-p-coumaroyl)sophoroside-5-(6-malonyl)glucoside A |
| P4 | 42.8 | 522 | 757 | 595(100),449(11),287(61) | Cyanidin-3-(6-p-coumaroyl)glucoside-5-glucoside A |
| P5 | 43.2 | 530 | 1081 | 919(15),449(6),287(6) | Cyanidin-3-(6-p-coumaroyl-6′-caffeoyl)-5-glucoside isomer1 A |
| P6 | 43.9 | 532 | 1167 | 919(44),757(5),287(20) | Cyanidin-3-(6-p-coumaroyl-6′-caffeoyl)sophoroside-5-(6-malonyl)glucoside isomer 1 A |
| P7 | 44.6 | 530 | 1167 | 919(27),757(5),287(6) | Cyanidin-3-(6 p-coumaroyl-6′-caffeoyl)sophoroside-5-(6-malonyl)glucoside isomer 2 A |
| P8 | 45.5 | 530 | 1081 | 919(100),449(11),287(20) | Cyanidin-3-(6-p-coumaroyl-6′-caffeoyl)sophoroside-5-glucoside isomer2 A |
| P9 | 45.9 | 530 | 1065 | 903(20),449(5),287(3) | Cyanidin-3-(6,6′-di p-coumaroyl)sophoroside-5-glucoside A |
| P10 | 46.4 | 526 | 1151 | 989(10),903(5),287(5) | Cyanidin-3-(6,6′-di p-coumaroyl)sophoroside-5-(6-malonyl)glucoside A |
| P11 | 47.0 | 514 | 1049 | 887(33),433(9),271(5) | Pelargonidin-3-(6,6′-di p-coumaroyl)sophoroside-5-glucoside B |
| P12 | 48.0 | 526 | 1167 | 1005(63),919(23),449(8),287(13) | Cyanidin-3-(6- p-coumaroyl-X-malonyl-6′-caffeoyl)sophoroside-5-glucoside A |
| P13 | 48.7 | 530 | 1151 | 989(28),449(17),287(5) | Cyanidin-3-(6- p-coumaroyl-X-malonyl-6′-p-coumaroyl)sophoroside-5-glucoside A |
| Five-Level CCCD Experimental Design | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Runs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
| Coded values | X1: Time (t) | −1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | 1.68 | −1.68 | 0 | 0 | 0 | 0 | −1.68 | −1.68 | −1.68 | −1.68 | 1.68 | 1.68 | 1.68 | 1.68 | 0 | 0 | 0 | 0 | 0 | 0 |
| X2: Temp. (T) | −1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 0 | 0 | −1.68 | 1.68 | 0 | 0 | −1.68 | −1.68 | 1.68 | 1.68 | −1.68 | −1.68 | 1.68 | 1.68 | 0 | 0 | 0 | 0 | 0 | 0 | |
| X3: Solvent (S) | −1 | 1 | −1 | 1 | −1 | 1 | −1 | 1 | 0 | 0 | 0 | 0 | −1.68 | 1.68 | −1.68 | 1.68 | −1.68 | 1.68 | −1.68 | 1.68 | −1.68 | 1.68 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Natural values | X1: t (min) | 40.3 | 40.3 | 40.3 | 40.3 | 99.7 | 99.7 | 99.7 | 99.7 | 120.0 | 20.0 | 70.0 | 70.0 | 70.0 | 70.0 | 20.0 | 20.0 | 20.0 | 20.0 | 120.0 | 120.0 | 120.0 | 120.0 | 70.0 | 70.0 | 70.0 | 70.0 | 70.0 | 70.0 |
| X2: T (°C) | 37.2 | 37.2 | 72.8 | 72.8 | 37.2 | 37.2 | 72.8 | 72.8 | 55.0 | 55.0 | 25.0 | 85.0 | 55.0 | 55.0 | 25.0 | 25.0 | 85.0 | 85.0 | 25.0 | 25.0 | 85.0 | 85.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | |
| X3: S (%) | 20.3 | 79.7 | 20.3 | 79.7 | 20.3 | 79.7 | 20.3 | 79.7 | 50.0 | 50.0 | 50.0 | 50.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | |
| Response Variables for RSM Application | |||||||||||||||||||||||||||||
| P1 | 3.34 | 5.61 | 4.50 | 5.75 | 3.41 | 5.80 | 3.55 | 4.96 | 3.93 | 5.08 | 5.05 | 5.52 | 3.17 | 5.78 | 1.96 | 6.36 | 4.56 | 5.83 | 2.27 | 5.86 | 1.42 | 5.11 | 4.93 | 4.91 | 5.34 | 5.35 | 5.24 | 4.84 | |
| P2 | 2.47 | 4.24 | 2.64 | 4.15 | 2.38 | 4.27 | 2.26 | 3.72 | 2.55 | 3.32 | 3.15 | 2.92 | 2.31 | 5.31 | 1.78 | 4.85 | 2.50 | 4.11 | 1.83 | 4.22 | 1.45 | 3.83 | 3.43 | 3.54 | 3.47 | 3.48 | 3.44 | 3.29 | |
| P3 | 3.94 | 5.53 | 4.71 | 5.37 | 3.60 | 6.52 | 3.90 | 5.78 | 3.93 | 4.89 | 5.29 | 5.74 | 2.81 | 5.13 | 2.10 | 4.25 | 2.98 | 3.16 | 1.59 | 4.63 | 1.48 | 5.42 | 6.33 | 7.04 | 6.70 | 6.71 | 6.94 | 6.67 | |
| P4 | 2.95 | 5.68 | 2.82 | 5.05 | 2.93 | 6.60 | 2.71 | 5.01 | 3.70 | 4.26 | 4.98 | 4.97 | 1.66 | 4.60 | 1.79 | 5.27 | 1.47 | 4.67 | 1.59 | 4.61 | 1.39 | 5.12 | 4.59 | 4.57 | 4.78 | 4.79 | 4.40 | 4.42 | |
| P5 | 7.61 | 13.13 | 8.39 | 13.66 | 7.30 | 13.00 | 8.15 | 13.66 | 10.54 | 12.68 | 15.27 | 17.09 | 2.64 | 9.81 | 1.87 | 8.32 | 1.47 | 10.19 | 1.59 | 8.67 | 1.39 | 11.07 | 16.99 | 16.40 | 17.62 | 17.66 | 15.93 | 16.40 | |
| P6 | 3.47 | 5.69 | 3.78 | 5.34 | 3.48 | 4.99 | 3.52 | 5.02 | 3.91 | 5.09 | 5.68 | 5.74 | 1.70 | 4.01 | 1.74 | 4.70 | 1.47 | 3.76 | 1.59 | 3.99 | 1.37 | 3.25 | 6.35 | 6.10 | 6.62 | 6.64 | 6.34 | 6.31 | |
| P7 | 2.52 | 4.25 | 2.62 | 3.98 | 2.43 | 3.59 | 2.41 | 3.62 | 3.12 | 3.52 | 3.80 | 3.64 | 1.75 | 4.07 | 1.74 | 4.83 | 1.91 | 3.98 | 1.59 | 4.07 | 1.37 | 3.28 | 3.73 | 3.75 | 3.85 | 3.86 | 3.69 | 3.69 | |
| P8 | 3.53 | 5.93 | 3.67 | 5.39 | 3.40 | 6.22 | 3.36 | 5.03 | 3.95 | 5.09 | 5.42 | 5.28 | 1.96 | 3.84 | 1.85 | 4.65 | 1.47 | 3.82 | 1.59 | 3.96 | 1.37 | 3.49 | 5.86 | 5.64 | 5.88 | 5.89 | 6.01 | 5.57 | |
| P9 | 5.10 | 8.37 | 5.33 | 7.82 | 4.78 | 8.55 | 4.94 | 7.82 | 5.46 | 7.15 | 8.25 | 8.59 | 2.07 | 5.44 | 1.94 | 5.27 | 1.47 | 4.98 | 1.59 | 4.70 | 1.40 | 5.41 | 9.99 | 9.52 | 10.25 | 10.27 | 9.49 | 9.71 | |
| P10 | 14.92 | 21.10 | 16.36 | 24.14 | 14.84 | 18.68 | 16.01 | 24.14 | 21.50 | 23.40 | 26.73 | 30.48 | 6.96 | 14.17 | 2.29 | 9.46 | 2.35 | 13.48 | 1.59 | 10.19 | 1.43 | 17.38 | 32.57 | 32.82 | 33.99 | 34.06 | 32.48 | 33.27 | |
| P11 | 10.18 | 14.08 | 13.19 | 15.07 | 9.34 | 12.26 | 10.90 | 13.27 | 12.83 | 17.10 | 16.97 | 19.23 | 5.98 | 5.71 | 2.31 | 5.13 | 5.67 | 5.26 | 1.27 | 4.82 | 1.16 | 5.32 | 20.04 | 19.79 | 20.91 | 20.95 | 21.22 | 19.94 | |
| P12 | 2.91 | 4.60 | 3.27 | 4.31 | 2.66 | 4.10 | 2.76 | 3.81 | 3.22 | 4.13 | 4.19 | 4.26 | 2.20 | 4.09 | 1.92 | 4.80 | 2.74 | 4.17 | 1.59 | 4.29 | 1.41 | 3.54 | 3.93 | 3.80 | 3.83 | 3.84 | 3.94 | 3.83 | |
| P13 | 4.84 | 7.26 | 5.77 | 6.99 | 4.30 | 7.53 | 4.89 | 6.72 | 5.63 | 7.31 | 7.30 | 7.85 | 2.73 | 4.57 | 2.08 | 4.75 | 3.32 | 4.12 | 1.59 | 4.08 | 1.40 | 4.15 | 8.06 | 8.08 | 8.34 | 8.36 | 8.75 | 8.38 | |
| TAC | 67.78 | 105.46 | 77.06 | 107.01 | 64.86 | 102.12 | 69.37 | 102.56 | 84.25 | 103.00 | 112.07 | 121.30 | 37.96 | 76.52 | 25.38 | 72.65 | 33.37 | 71.53 | 21.22 | 68.10 | 18.04 | 76.37 | 116.79 | 115.97 | 111.59 | 111.85 | 117.87 | 116.32 | |
| Yield | 36.35 | 28.58 | 38.26 | 31.41 | 36.08 | 29.95 | 39.41 | 32.14 | 38.12 | 37.53 | 34.75 | 37.90 | 38.84 | 18.08 | 35.62 | 13.22 | 38.27 | 17.80 | 35.42 | 16.52 | 41.00 | 20.24 | 35.68 | 34.54 | 35.68 | 35.61 | 35.54 | 35.40 | |
| Response variables | Fitting Coefficients Obtained after Applying the Second-Order Polynomial Equation with Interactive Terms | Optimal Processing Conditions and Response Values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept | Linear Effect | Quadratic Effect | Interactive Effect | R2 | t (min) | T (°C) | S (%) | Optimum | |||||||
| b0 | b1 (t) | b2 (T) | b3 (S) | b11 (t2) | b22 (T2) | b33 (S2) | b12 (tT) | b13 (tS) | b23 (TS) | ||||||
| P1 | 5.06 ± 0.15 | −0.28 ± 0.09 | ns | 0.92 ± 0.09 | −0.16 ± 0.11 | ns | −0.17 ± 0.11 | −0.17 ± 0.06 | 0.07 ± 0.06 | −0.15 ± 0.06 | 0.9441 | 81.06 ± 2.08 | 20.00 ± 1.73 | 100.00 ± 1.58 | 6.56 ± 0.31 |
| P2 | 3.37 ± 0.10 | −0.15 ± 0.06 | ns | 0.76 ± 0.06 | −0.20 ± 0.07 | ns | 0.11 ± 0.07 | −0.04 ± 0.00 | ns | −0.07 ± 0.04 | 0.9556 | 64.04 ± 5.07 | 20.00 ± 0.43 | 100.00 ± 9.11 | 5.15 ± 0.36 |
| P3 | 31.23 ± 1.68 | ns | 0.96 ± 0.95 | ns | −2.14 ± 1.15 | ns | −5.40 ± 1.15 | ns | 0.98 ± 0.68 | ns | 0.9359 | 70.00 ± 3.94 | 90.00 ± 6.07 | 50.00 ± 3.80 | 32.85 ± 2.47 |
| P4 | 4.65 ± 0.19 | ns | −0.20 ± 0.11 | 1.06 ± 0.11 | −0.11 ± 0.01 | ns | −0.41 ± 0.13 | ns | ns | ns | 0.9225 | 70.00 ± 5.49 | 20.00 ± 1.94 | 88.44 ± 5.39 | 5.67 ± 1.05 |
| P5 | 15.59 ± 0.62 | ns | 0.33 ± 0.22 | 2.42 ± 0.35 | −0.95 ± 0.42 | ns | −2.85 ± 0.42 | ns | ns | ns | 0.9449 | 70.00 ± 1.26 | 90.00 ± 5.25 | 62.60 ± 5.86 | 16.66 ± 1.76 |
| P6 | 5.90 ± 0.27 | −0.16 ± 0.15 | −0.10 ± 0.01 | 0.74 ± 0.15 | −0.31 ± 0.18 | ns | −0.89 ± 0.18 | ns | ns | ns | 0.9336 | 62.47 ± 1.24 | 20.00 ± 0.31 | 62.27 ± 3.93 | 6.24 ± 0.56 |
| P7 | 3.63 ± 0.08 | −0.16 ± 0.05 | −0.09 ± 0.05 | 0.70 ± 0.05 | −0.08 ± 0.06 | ns | −0.23 ± 0.06 | ns | ns | −0.07 ± 0.03 | 0.9701 | 40.93 ± 1.60 | 20.00 ± 1.49 | 100.00 ± 6.12 | 4.59 ± 0.34 |
| P8 | 5.59 ± 0.18 | −0.13 ± 0.10 | −0.14 ± 0.10 | 0.77 ± 0.10 | −0.23 ± 0.12 | ns | −0.80 ± 0.12 | ns | ns | ns | 0.9456 | 61.55 ± 4.64 | 20.00 ± 1.03 | 64.30 ± 1.89 | 6.03 ± 0.55 |
| P9 | 9.06 ± 0.43 | ns | −0.12 ± 0.02 | 1.15 ± 0.24 | −0.62 ± 0.30 | ns | −1.52 ± 0.30 | ns | ns | ns | 0.9286 | 70.00 ± 1.16 | 20.00 ± 1.24 | 61.18 ± 2.85 | 9.47 ± 1.32 |
| P10 | 29.87 ± 1.32 | ns | 0.85 ± 0.74 | 2.89 ± 0.74 | −2.07 ± 0.91 | ns | −6.27 ± 0.91 | ns | ns | ns | 0.9377 | 70.00 ± 1.42 | 90.00 ± 2.46 | 56.85 ± 0.94 | 31.63 ± 2.42 |
| P11 | 8.93 ± 0.25 | −0.32 ± 0.14 | −0.12 ± 0.14 | 0.63 ± 0.14 | −0.47 ± 0.17 | ns | −1.64 ± 0.17 | ns | ns | −0.10 ± 0.01 | 0.9359 | 59.99 ± 5.19 | 20.00 ± 0.59 | 57.18 ± 0.16 | 9.28 ± 0.38 |
| P12 | 3.91 ± 0.06 | −0.22 ± 0.03 | −0.03 ± 0.03 | 0.65 ± 0.03 | −0.06 ± 0.04 | ns | −0.25 ± 0.04 | −0.05 ± 0.02 | ns | −0.09 ± 0.02 | 0.9757 | 36.49 ± 0.30 | 20.00 ± 1.21 | 98.54 ± 9.29 | 4.71 ± 0.20 |
| P13 | 7.93 ± 0.20 | −0.26 ± 0.11 | −0.16 ± 0.11 | 0.73 ± 0.11 | −0.37 ± 0.03 | ns | −1.37 ± 0.13 | ns | 0.09 ± 0.08 | −0.10 ± 0.01 | 0.9451 | 60.74 ± 0.00 | 20.00 ± 1.30 | 59.46 ± 0.04 | 8.38 ± 0.29 |
| TAC | 109.78 ± 2.73 | −1.93 ± 1.54 | 1.07 ± 0.32 | 14.30 ± 1.54 | −6.20 ± 1.87 | ns | −17.00 ± 1.87 | ns | ns | ns | 0.9577 | 65.37 ± 3.62 | 90.00 ± 1.17 | 62.50 ± 4.24 | 114.74 ± 0.58 |
| Yield | 36.43 ± 1.46 | 0.49 ± 0.88 | 1.19 ± 0.87 | −5.56 ± 0.87 | ns | ns | −3.09 ± 0.84 | ns | ns | ns | 0.9592 | 120.00 ± 2.62 | 90.00 ± 7.72 | 23.23 ± 0.91 | 41.77 ± 1.59 |
| Anthocyanin Compounds | Equations | Equation Numbers |
|---|---|---|
| P1 | Equation (2) | |
| P2 | Equation (3) | |
| P3 | Equation (4) | |
| P4 | Equation (5) | |
| P5 | Equation (6) | |
| P6 | Equation (7) | |
| P7 | Equation (8) | |
| P8 | Equation (9) | |
| P9 | Equation (10) | |
| P10 | Equation (11) | |
| P11 | Equation (12) | |
| P12 | Equation (13) | |
| P13 | Equation (14) | |
| TAC | Equation (15) | |
| Yield | Equation (16) |
| (A) Maximum Response Values (mg/g of Extract) of the Individual Anthocyanin Compounds | |||||||||||||
| Peak: | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 |
| Optimum: | 6.56 | 5.15 | 32.85 | 5.67 | 16.66 | 6.24 | 4.59 | 6.03 | 9.47 | 31.63 | 9.28 | 4.71 | 8.38 |
| (B) Values of each Anthocyanin Compound at the Optimal Conditions of the other Compounds | |||||||||||||
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | |
| P1 | 1 | 0.99 | 0.77 | 0.96 | 0.81 | 0.84 | 0.95 | 0.85 | 0.83 | 0.79 | 0.81 | 0.94 | 0.82 |
| P2 | 0.99 | 1 | 0.65 | 0.91 | 0.71 | 0.73 | 0.98 | 0.74 | 0.72 | 0.68 | 0.70 | 0.95 | 0.71 |
| P3 | 0.45 | 0.42 | 1 | 0.63 | 0.97 | 0.87 | 0.33 | 0.85 | 0.88 | 0.99 | 0.88 | 0.33 | 0.88 |
| P4 | 0.99 | 0.99 | 0.76 | 1 | 0.83 | 0.94 | 0.97 | 0.95 | 0.94 | 0.80 | 0.92 | 0.97 | 0.93 |
| P5 | 0.65 | 0.66 | 0.97 | 0.80 | 1 | 0.93 | 0.61 | 0.93 | 0.93 | 0.99 | 0.92 | 0.61 | 0.93 |
| P6 | 0.75 | 0.77 | 0.92 | 0.89 | 0.94 | 1 | 0.74 | 1.00 | 1.00 | 0.94 | 1.00 | 0.75 | 1.00 |
| P7 | 0.97 | 0.99 | 0.76 | 0.97 | 0.80 | 0.90 | 1 | 0.91 | 0.88 | 0.79 | 0.87 | 1.00 | 0.88 |
| P8 | 0.79 | 0.81 | 0.89 | 0.91 | 0.92 | 1.00 | 0.79 | 1 | 1.00 | 0.91 | 0.99 | 0.80 | 1.00 |
| P9 | 0.72 | 0.72 | 0.93 | 0.86 | 0.96 | 1.00 | 0.66 | 0.99 | 1 | 0.95 | 0.99 | 0.66 | 0.99 |
| P10 | 0.48 | 0.49 | 0.99 | 0.69 | 0.99 | 0.90 | 0.43 | 0.89 | 0.91 | 1 | 0.90 | 0.44 | 0.90 |
| P11 | 0.61 | 0.63 | 0.94 | 0.80 | 0.93 | 0.99 | 0.61 | 0.99 | 0.99 | 0.94 | 1 | 0.63 | 1.00 |
| P12 | 0.97 | 0.99 | 0.82 | 0.98 | 0.85 | 0.91 | 1.00 | 0.92 | 0.90 | 0.84 | 0.89 | 1 | 0.90 |
| P13 | 0.69 | 0.70 | 0.91 | 0.84 | 0.91 | 1.00 | 0.67 | 1.00 | 1.00 | 0.92 | 1.00 | 0.68 | 1 |
| Quantification (mg/g E) | L* | a* | b* | Conversion Color to RGB Values |
|---|---|---|---|---|
| 115.4 ± 0.4 | 20.5 ± 0.5 | 33.0 ± 0.1 | 8.2 ± 0.4 |
| Antibacterial Activity | |||||||
| B.c. | S.a. | L.m. | E.c. | En.cl. | S.t. | ||
| Anthocyanins rich extract | MIC | 0.037 | 0.075 | 0.05 | 0.037 | 0.075 | 0.15 |
| MBC | 0.075 | 0.15 | 0.075 | 0.075 | 0.15 | 0.30 | |
| Streptomycin (1) | MIC | 0.10 | 0.04 | 0.20 | 0.20 | 0.20 | 0.20 |
| MBC | 0.20 | 0.10 | 0.30 | 0.30 | 0.30 | 0.30 | |
| Ampicillin (1) | MIC | 0.25 | 0.25 | 0.40 | 0.40 | 0.25 | 0.75 |
| MBC | 0.40 | 0.45 | 0.50 | 0.50 | 0.50 | 1.20 | |
| Antifungal Activity | |||||||
| A.fun. | A.o. | A.n. | P.f. | P.o | P.v.c. | ||
| Anthocyanins rich extract | MIC | 0.037 | 0.002 | 0.075 | 0.075 | 0.30 | 0.30 |
| MFC | 0.075 | 0.075 | 0.15 | 0.15 | 0.45 | 0.45 | |
| Ketoconazole (1) | MIC | 0.25 | 0.20 | 0.20 | 0.20 | 2.50 | 0.20 |
| MFC | 0.50 | 0.50 | 0.50 | 0.50 | 3.50 | 0.30 | |
| Bifonazole (1) | MIC | 0.15 | 0.10 | 0.15 | 0.20 | 0.20 | 0.10 |
| MFC | 0.20 | 0.20 | 0.20 | 0.25 | 0.25 | 0.20 | |
| Tumor Cell Lines | Concentrations (GI50 Values, µg/mL) |
|---|---|
| MCF-7 (breast carcinoma) | >400 |
| NCI-H460 (lung carcinoma) | >400 |
| HeLa (cervical carcinoma) | 213 ± 9 |
| HepG2 (hepatocellular carcinoma) | 198 ± 9 |
| Non-Tumour Cells | |
| PLP2 (non-tumor porcine liver primary cells) | >400 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, F.; Pereira, E.; Prieto, M.A.; Calhelha, R.C.; Ćirić, A.; Soković, M.; Simal-Gandara, J.; Barros, L.; Ferreira, I.C.F.R. Optimization of the Extraction Process to Obtain a Colorant Ingredient from Leaves of Ocimum basilicum var. purpurascens. Molecules 2019, 24, 686. https://doi.org/10.3390/molecules24040686
Fernandes F, Pereira E, Prieto MA, Calhelha RC, Ćirić A, Soković M, Simal-Gandara J, Barros L, Ferreira ICFR. Optimization of the Extraction Process to Obtain a Colorant Ingredient from Leaves of Ocimum basilicum var. purpurascens. Molecules. 2019; 24(4):686. https://doi.org/10.3390/molecules24040686
Chicago/Turabian StyleFernandes, Filipa, Eliana Pereira, Miguel A. Prieto, Ricardo C. Calhelha, Ana Ćirić, Marina Soković, Jesus Simal-Gandara, Lillian Barros, and Isabel C. F. R. Ferreira. 2019. "Optimization of the Extraction Process to Obtain a Colorant Ingredient from Leaves of Ocimum basilicum var. purpurascens" Molecules 24, no. 4: 686. https://doi.org/10.3390/molecules24040686