Abstract
Mass spectrometry (MS)-based, integrated proteomics, metabolomics, and lipidomics (collectively, multi-omics) studies provide a very detailed snapshot of virus-induced changes to the host following infection and can lead to the identification of novel prophylactic and therapeutic targets for preventing or lessening disease severity. Multi-omics studies with Middle East respiratory syndrome coronavirus (MERS-CoV) are challenging as the requirements of biosafety level 3 containment limit the numbers of samples that can be safely managed. To address these issues, the multi-omics sample preparation technique MPLEx (metabolite, protein, and lipid extraction) was developed to partition a single sample into three distinct parts (metabolites, proteins, and lipids) for multi-omics analysis, while simultaneously inactivating MERS-CoV by solubilizing and disrupting the viral envelope and denaturing viral proteins. Here we describe the MPLEx protocol, highlight the step of inactivation, and describe the details of downstream processing, instrumental analysis of the three separate analytes, and their subsequent informatics pipelines.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- MPLEx
- Metabolomics
- Proteomics
- Lipidomics
- MERS-CoV
- Virus-host interactions
- Mass spectrometry (MS)
- Virus inactivation
1 Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) research is of global interest due to the high case fatality rate, narrowly defined epidemiology, and spread of the virus to 27 countries to date. The World Health Organization (WHO) recognizes the urgent need for effective public health countermeasures due to the ongoing epidemic, and MERS-CoV is on the WHO list of priority pathogens to highlight the critical need for the development of diagnostic products and prophylactic and therapeutic treatment options [1]. The National Institute of Allergy and Infectious Diseases Systems Biology for Infectious Disease Research Program was established to support research focusing on multi-omics approaches and dataset integration to develop and validate predictive models of infectious disease initiation, progression, and outcomes [2]. Thus, there is great interest and need for improved understanding of the pathogenesis of MERS-CoV; however, challenges remain in effective study of this pathogen.
For example, samples from MERS-CoV infected patients are virtually unavailable for analysis, and animal models that recapitulate disease phenotypes seen in humans have only recently been generated, both of which have drastically slowed our progress toward understanding MERS-CoV pathogenesis in the host [3]. Systems biology studies offer a way to capture big picture snapshots of individual cellular components (proteins, lipids, metabolites) that are modulated over the course of infection to develop a better understanding of pathogen-host interactions [4,5,6,7,8,9,10,11,12,13].
Proteins are the major effectors of cellular pathways and represent the dynamic expression of information encoded within the genome during infection. Protein driven cellular responses following infection can favor either viral clearance or spread; therefore, taking snapshots of total proteins isolated from infected cells over the course of infection can provide insights into their underlying molecular mechanisms of pathogenicity, and potentially even single out targets for pharmacological intervention [14].
Metabolites are biomolecules required for cellular metabolism and can either be intermediates produced during cellular metabolic processes or end products of cellular pathways. They represent the level of homeostasis of cellular activities in a host [15, 16]. Importantly, certain metabolites play key roles during the cellular responses to various viral infections such as signaling, initiating or resolving inflammation, or other immune related responses [17]. Therefore, metabolite levels can be profiled between healthy and disease states to not only understand the triggers of change but to also discover possible biomarkers in early disease stages.
Lipids have key functions in signaling pathways, energy storage, and the structural integrity of cell membranes. They also function in host-pathogen interactions and immunomodulation since they act in first-line recognition and host cell signaling during pathogen docking, invasion, and intracellular trafficking [18]. Lipid metabolism and cellular lipids are greatly affected by virus infections by inducing major lipid modifications within host cells through the production of convoluted membranes and double membrane vesicles (DMVs) [19,20,21,22]. Virus-induced production of membrane networks and organelles is a common occurrence among all positive sense RNA viruses [23, 24]. The roles of these virus-induced DMVs are not fully understood; however, evidence suggests some viruses may use them for replication, to conceal viral RNA from host antiviral response, or they may have roles in autophagy as autophagosomes [25].
While it’s clear that metabolites, proteins, and lipids play an important role in fully characterizing the MERS-CoV infection, the proteomic, metabolomic, and lipidomic sample manipulation of MERS-CoV outside of appropriate biosafety level (BSL) containment laboratories can take place only subsequent to pathogen inactivation. Here, we describe the MPLEx (metabolite, protein, and lipid extraction) protocol for the extraction of protein, metabolites, and lipids from a single sample that simultaneously inactivates the MERS-CoV virus [5, 6, 26]. Each analyte can then be analyzed by the respective mass spectrometry (MS)-based omics pipeline.
To illustrate the effectiveness of this protocol, Nakayasu et al. performed an integrative multi-omics study using a human lung epithelial cell line infected with MERS-CoV, which showed the impact of the viral infection on the host glycolytic pathway, different host metabolic pathways, and also global changes in lipid profiles induced by infection [5]. To illustrate the effectiveness of MPLEx on pathogen inactivation, Burnum-Johnson et al. showed complete inactivation of both bacterial and viral pathogens with exposed lipid envelopes, including MERS-CoV [6].
The MPLEx method is a simple yet powerful protocol that can be applied for integrative multi-omic measurements while concurrently inactivating MERS-CoV (or other enveloped viruses). The multiple analyte samples obtained from MPLEx can be used across various instrument and data analysis platforms. Here we describe the pipeline as implemented at Pacific Northwest National Laboratory.
2 Materials
2.1 Extraction Chemicals, Quality Controls, and Mobile Phases
-
1.
18.2 MΩ cm water (see Note 1).
-
2.
Ammonium bicarbonate buffer (NH4HCO3): 150 mM, pH 8.0, ice cold.
-
3.
Rapid Quench Buffer: 60% methanol, 0.85% ammonium bicarbonate in sterile water. Store at −80 °C and stand in biological safety cabinet to warm up during processing.
-
4.
MPLEx Solution: two volumes chloroform to one volume methanol (2:1, v/v). Store at −80 °C and stand in biological safety cabinet to warm up to −20 °C during processing (see Note 2).
-
5.
High-performance liquid chromatography (HPLC) solvent A: 10 mM ammonium formate, pH 10.0.
-
6.
HPLC solvent B: 10:90 10 mM ammonium formate, pH 10.0:acetonitrile (ACN).
-
7.
Proteomics MS mobile phase (MP) A: 0.1% formic acid in water.
-
8.
Proteomics MS MP B: 0.1% formic acid in ACN.
-
9.
Lipidomics MS MP A: ACN:H2O (40:60) in 10 mM ammonium acetate.
-
10.
Lipidomics MS MP B: ACN:isopropyl alcohol (10:90) in 10 mM ammonium acetate.
-
11.
Metabolomics MS MP: Helium gas.
-
12.
Metabolomics retention time alignment compounds: FAMEs (fatty acid methyl esters) (Agilent, Santa Clara, CA) [27].
-
13.
Lipid quality control: Bovine Brain Total Lipid Extract (BTLE) (Avanti Polar Lipids, Inc., Alabaster, AL).
2.2 Metabolite Derivatization Chemicals
-
1.
Anhydrous pyridine (99.8% purity) (see Note 3).
-
2.
O-Methylhydroxylamine hydrochloride (>98% purity).
-
3.
Silylation Reagent: N-Methyl-N-(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane (MSTFA with 1% of TMCS) (GC grade) (see Note 4).
2.3 Tryptic Digestion, Solid Phase Extraction, Tandem Mass Tag (TMT) Isobaric Labeling, and High-Performance Liquid Chromatography (HPLC) Fractionation Chemicals
-
1.
NH4HCO3 buffer : 100 mM, pH ~8.4 (see Note 5).
-
2.
Bicinchoninic acid (BCA) assay reagent kit (Thermo Scientific, Waltham, MA).
-
3.
Urea protein denaturing solution: 8 M in 100 mM NH4HCO3, pH ~8.0.
-
4.
Protein-reducing solution: 0.5 M dithiothreitol (DTT) in 100 mM NH4HCO3, pH 8.4.
-
5.
Protein alkylation solution: 0.4 M iodoacetamide (IAM) in 100 mM NH4HCO3, pH 8.4.
-
6.
1 M CaCl2 in 100 mM NH4HCO3, pH 8.4.
-
7.
Sequencing-grade modified trypsin (see Note 6).
-
8.
C-18 Solid Phase Extraction (SPE) column conditioning 1: methanol.
-
9.
C-18 SPE column conditioning 2: 0.1% trifluoroacetic acid (TFA) in H2O.
-
10.
C-18 SPE column washing: 95:5H2O:ACN, 0.1% TFA.
-
11.
C-18 SPE column elution: 80:20 ACN:H2O, 0.1% TFA.
-
12.
Amine-reactive Thermo Scientific Tandem Mass Tag (TMT) Isobaric Mass Tagging Kits (Thermo Scientific, Rockford, IL).
-
13.
TMT resuspension reagent: Anhydrous ACN.
-
14.
TMT dissolution buffer: 50 mM triethylammonium bicarbonate (TEAB), pH 8.5.
-
15.
TMT reaction quenching buffer: 5% hydroxylamine.
2.4 Personal Protective Equipment (PPE) and Lab Supplies
-
1.
Milli-Q water purification system.
-
2.
Vortex.
-
3.
Refrigerated centrifuge.
-
4.
Thermomixer with Thermotop.
-
5.
Magnetic stir plate and bars.
-
6.
pH paper strips, pH range 0–14.
-
7.
Breathe Easier plate membranes.
-
8.
−20 °C freezer.
-
9.
−80 °C freezer.
-
10.
Conical glass vials for lipids.
-
11.
Glass vial lids without pre-slit septa.
-
12.
Glass MS sample vial and inserts for proteomics.
-
13.
Pipette set (2–1000 μL).
-
14.
Kimwipes.
-
15.
Chloroform-resistant polypropylene pipette tips (see Note 7).
-
16.
Chloroform compatible 1.7 mL or 2 mL Sorenson MμlTI™ SafeSeal™ Microcentrifuge Tubes (see Note 8).
-
17.
Hamilton Glass Syringes, 5 mL, 50 μL, and 250 μL.
-
18.
Bath Sonicator.
-
19.
C-18 solid phase extraction columns 50 mg/1 mL.
-
20.
Liquid nitrogen.
-
21.
Speedvac Concentrator (for drying in vacuo).
-
22.
Biocontainment rotor Eppendorf centrifuge.
-
23.
Serological pipette.
-
24.
Gilson pipetman and non-aerosol tips (Gilson, Middleton, WI).
-
25.
3 M Powered air-purifying respirator.
-
26.
3 M Versaflo hood with shroud.
-
27.
3 M Heavy-duty rubber breathing tube.
-
28.
3 M Belt for PAPR.
-
29.
3 M Cartridge filer.
-
30.
3 M Filter cover.
-
31.
3 M Battery.
-
32.
3 M Battery charger.
-
33.
3 M Airflow indicator.
-
34.
DuPont Tyvek IsoClean Coveralls.
-
35.
DuPont ProClean Boot Covers.
-
36.
Kappler ProVent 7000 Isolation Gowns.
-
37.
Cardinal Health Esteem Nitrile Gloves with Neu-Thera.
-
38.
70% Ethanol (surface decontamination).
-
39.
CiDecon™ Disinfectant—Concentrated phenolic disinfectant.
-
40.
Autoclave (Steris).
2.5 Instrumentation, Columns, and Software
-
1.
Microplate reader (Epoch).
-
2.
Gilson GX-274 ASPEC™ 4-probe positive pressure automated SPE system with 406 Dual Syringe Pumps (Gilson, Middleton, WI).
-
3.
Proteomics off-line fractionation: Agilent 1200 HPLC System equipped with a quaternary pump, degasser, diode array detector, peltier-cooled autosampler, and fraction collector (set at 4 °C) (Agilent, Santa Clara, CA).
-
4.
Proteomics off-line fractionation column: XBridge C-18 reversed-phase (RP) HPLC column, 250 mm × 4.6 mm, containing 5-μm particles, and a 4.6 mm × 20 mm guard column (Waters, Milford, MA).
-
5.
Proteomics LC-MS: M-Class nanoAcquity dual pumping UPLC (Waters, Milford, MA) coupled with a Q Exactive HF Hybrid Quadrupole-Orbitrap MS (Thermo Scientific, San Jose, CA).
-
6.
Proteomics LC column: Jupiter 3 μm and 5 μm C-18 media (Phenomenex, Torrance, CA), a 70-cm length of 360 μm o.d. × 75 μm i.d. and 4-cm length of 360 μm o.d. × 150 μm i.d. of fused silica (Molex, Lisle, IL).
-
7.
Proteomics MS additional part 1: Kasil-based frits (Next Advance, Inc., Troy, NY).
-
8.
Proteomics MS additional part 2: Nanospray Flex Ion Source (Thermo Scientific, San Jose, CA).
-
9.
Metabolomics Gas Chromatography MS (GC-MS): Agilent 7890A GC coupled with a single quadrupole 5975C MS (Agilent Technologies, Inc., Santa Clara, CA).
-
10.
Metabolomics GC column: HP-5MS or DB-5MS GC capillary column 30 m × 0.25 mm × 0.25 μm (Agilent, Santa Clara, CA).
-
11.
Lipidomics LC-MS: H-Class nanoAcquity dual pumping UPLC (Waters, Milford, MA) coupled with a Velos Pro Orbitrap MS (Thermo Scientific, San Jose, CA).
-
12.
Lipidomics LC column: Analytical LC column CSH 3.0 mm × 150 mm × 1.7 μm particle size (Waters, Milford, MA).
-
13.
Proteomics software 1: MSCovert for spectrum to peak list conversion [28].
-
14.
Proteomics software 2: mzRefinery for mass recalibration [29].
-
15.
Proteomics software 3: MS-GF+ for peptide/protein identification [30].
-
16.
Proteomics software 4: MASIC for extracting intensity values of TMT reporter ions [31].
-
17.
Metabolomics software: MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC-MS-based metabolome analysis [32].
-
18.
Lipidomics software 1: LIQUID: an open-source software for identifying lipids [33].
-
19.
Lipidomics software 2: MZmine: framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data [34].
3 Methods
MERS-CoV-infected cells are first lysed and extracted into metabolites, proteins, and lipids using MPLEx under BSL3 containment. The three analytes (in separate phases) are then individually collected into different tubes and vials, and this is the point of pathogen inactivation. The separate tubes are then decontaminated and brought out of the biological safety cabinet.
The protein phase is tryptically digested, desalted with C-18 columns, TMT labeled (and combined), separated via off-line C-18 RPLC (reversed-phase LC), concatenated, and analyzed by an M-Class nanoAcquity dual pumping UPLC coupled with a Q Exactive HF Hybrid Quadrupole-Orbitrap MS. The subsequent datasets are analyzed using MSCovert for spectrum to peak list conversion, mzRefinery for mass recalibration, and MS-GF+ for peptide/protein identification [28,29,30]. Quantitative information is extracted from the TMT reporter ion intensities using MASIC [31].
The metabolite phase is dried in vacuo, derivatized, and analyzed via GC-MS, and detected peaks are matched against an appropriate library to make metabolite identifications.
The lipid phase is dried in vacuo, reconstituted in methanol, and analyzed via a Waters Acquity UPLC H-class system and Velos Pro Orbitrap mass spectrometer, and lipid identifications are made via the LIQUID software [33]. MZmine is then used to process, visualize, and analyze peaks corresponding to identified lipids [34].
3.1 MPLEx Sample Extraction Under BSL3 Conditions
All work at BSL3 must be performed within a certified biological safety cabinet in a room under negative pressure with dedicated supply and exhaust using all appropriate personal protective equipment (PPE).
-
1.
BSL3 Lysis: For each well in a 6-well plate, remove the media (disposing into a tray filled half way with 2× CiDecon™ phenolic disinfectant solution) and rinse cells with 3 mL of ~−40 °C Rapid Quenching Solution. Remove as quickly as possible and discard as above.
-
2.
Add 150 μL of ice-cold 150 mM ammonium bicarbonate buffer to each well and scrape cells from plate. Rinse well with buffer to make sure all cells are collected. Remove to fresh 1.7 mL Sorenson tube (see Notes 8 and 9).
-
3.
Add 600 μL −80 °C 2:1 chloroform/methanol (v/v) solution (fourfold excess) and close the tube tightly.
-
4.
Shake sample vigorously for 10 s.
-
5.
Incubate on ice for 5 min.
-
6.
Shake sample vigorously for 10 s (see Note 10). This is the stage of pathogen inactivation.
-
7.
Centrifuge at 13,000 × g for 10 min (in a centrifuge with a biocontainment lid) to separate the three phases (see Fig. 1).
-
8.
Remove the upper phase to a fresh 1.7 mL Sorenson tube that has been labeled with the same sample name and “metabolites.”
-
9.
With a fresh tip, go through the protein “disc” at the interface, lightly push out any protein that may have entered the tip and bring the tip to the bottom of the tube to collect the organic phase. Stop collecting well before you reach the interface. Place the sample into a conical glass vial that has been labeled with the same sample name and “lipids.”
-
10.
Freeze aqueous and organic phases to bring out of BSL3 containment.
-
11.
Add 200 μL 100% methanol to the protein “disc” and shake vigorously to mix. Pellet gently at 9000 × g for 5 min.
-
12.
Gently decant the supernatant into waste and allow the protein pellet to “semi” dry upside down standing on a Kimwipe in the biological safety cabinet for 5 min.
-
13.
Freeze at −80 °C prior to removing from BSL3.
-
14.
Surface decontaminate all samples with 70% ethanol, seal into secondary containment and remove from facility. Return to −80 °C freezer at BSL2.
-
15.
Dry in vacuo all aqueous phase and organic phase samples. Open tube and vial lids and place samples into slots. Use a no-heat setting and check progress after 2.5 h. Remove desiccated samples and replace with remaining ones until all samples have been dried and return desiccated samples to −80 °C prior to either shipping or further analysis (see Note 11).
-
16.
The lightly dried protein pellet is stored at −80 °C until processing (see Subheadings 3.2, 3.5, and 3.7).
-
17.
The dry metabolite samples are stored at −20 °C until processing (see Subheadings 3.3, 3.4, and 3.8).
-
18.
The dry lipid samples are reconstituted in 500 μL of 2:1 chloroform:methanol and stored at −20 °C with lids that do not have a pre-slit septa (to reduce evaporation) until processing (see Subheadings 3.6 and 3.9).
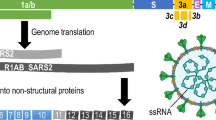
Schematic of the MPLEx phase separation and pathogen inactivation. Repurposed from Ref. [6] with permission from The Royal Society of Chemistry
3.2 Protein Tryptic Digestion, TMT Isobaric Labeling and High-Performance Liquid Chromatography (HPLC) Fractionation
-
1.
Denature and Reduce Protein:
-
(a)
Add up to 200 μL of 8 M urea solution to the protein pellet for protein denaturation.
-
(b)
Using the BCA reagent kit, perform a protein assay according to the manufacturer’s instructions. The protein assay is used to determine the initial protein mass of the sample prior to digestion (see Note 12).
-
(c)
Add enough 0.5 M DTT to get a final concentration of 5 mM for protein reduction.
-
(d)
Vortex and lightly water-bath sonicate the protein into solution.
-
(e)
Incubate at 60 °C for 30 min on a Thermomixer with a ThermoTop at 1000 rpm constant shaking.
-
(a)
-
2.
Alkylate:
-
(a)
Add enough 0.4 M IAM to each sample to get a final concentration of 40 mM.
-
(b)
Incubate for 1 h at room temperature, protected from light, on a Thermomixer with a ThermoTop at 1000 rpm constant shaking.
-
(a)
-
3.
Digest:
-
(a)
Dilute each sample tenfold with NH4HCO3, pH 8.4.
-
(b)
Add enough 1 M CaCl2 to reach a final concentration of 1 mM.
-
(c)
Incubate 1 μg/μL trypsin for 15 min at 37 °C to activate.
-
(d)
Add trypsin to each sample at a 1:50 (w/w) enzyme-to-protein ratio and incubate at 37 °C for 3 h on a Thermomixer with a ThermoTop at 700 rpm constant shaking.
-
(e)
Flash freeze with liquid nitrogen and store at −80 °C until solid phase extraction (SPE) desalting can be performed.
-
(a)
-
4.
C-18 SPE Desalting:
-
(a)
Thaw the sample and centrifuge at 10,000 × g for ~10 min to remove any precipitate and transfer to a fresh tube making sure not to pull up anything that might have pelleted.
-
(b)
Use one 50 mg/1 mL Supelco C-18 column per sample (capacity = 2.5 mg). C-18 SPE desalting can be performed manually on a vacuum manifold or using an automated SPE system.
-
Condition: 3× 1 mL methanol.
-
Equilibrate: 3× 1 mL 0.1% TFA.
-
Pass sample over column.
-
Wash: 4× 1 mL 95:5H2O:ACN, 0.1% TFA.
-
Allow column to go to dryness.
-
Elute: 1× 1 mL 80:20 ACN:H2O, 0.1% TFA into clean 1.5-mL microcentrifuge tube.
-
-
(c)
Concentrate in vacuo to approximately 50 μL (1.5–2 h).
-
(d)
Centrifuge at 10,000 × g for 2 min, perform BCA assay, and determine volume.
-
(e)
Aliquot an equal mass of peptide (30–100 μg each) into separate tubes and dry down the peptides (see Note 13).
-
(f)
Add 30 μL of the TMT dissolution buffer to each sample and ensure the pH is between 7 and 9 using pH paper.
-
(a)
-
5.
TMT Labeling:
-
(a)
Post-digestion TMT labeling is performed according to the manufacturer’s instructions.
-
(b)
Bring each needed vial of TMT Reagent to room temperature and spin to collect the liquid at the bottom.
-
(c)
Add 41 μL of TMT resuspension reagent to each TMT label, vortex, and spin. Allow to dissolve for 5 min with occasional vortexing.
-
(d)
Add each TMT label into each sample.
-
(e)
Incubate for 1 h at room temperature.
-
(f)
Add 8 μL of TMT reaction quenching buffer to the sample and incubate for 15 min to quench the reaction.
-
(g)
Combine all of the samples per set together and dry down to ~100 μL to remove the ACN.
-
(h)
Perform C-18 SPE clean up in order to remove excess tag from the sample as described previously (see Subheading 3.2, step 4a–d).
-
(i)
Pipette the sample into an HPLC vial.
-
(a)
-
6.
Reversed-Phase High-Performance Liquid Chromatography (RPLC):
The TMT-labeled sample is separated on a Waters reversed-phase XBridge C-18 column (250 mm × 4.6 mm column containing 5-μm particles, and a 4.6 mm × 20 mm guard column) using an Agilent 1200 HPLC System.
-
(a)
Reconstitute the sample up to 930 μL with HPLC Solvent A and inject onto the column at a flow rate of 0.5 mL/min.
-
(b)
After sample loading, the C-18 column is washed for 35 min with solvent A, before applying a 90-min LC gradient with solvent B. The LC gradient starts with a linear increase of solvent B to 10% in 10 min, then a linear increase to 20% B in 15 min, and 30 min to 30% B, 15 min to 35% B, 10 min to 45% B, and another 10 min to 100% solvent B. The flow rate is 0.5 mL/min [35].
-
(c)
Without collecting the first 15 min, a total of 96 fractions are collected into a 96-well plate throughout the LC gradient in equal time intervals.
-
(d)
At this point the plate can be frozen or dried down in vacuo.
-
(e)
After the fractions are dried, each fraction is reconstituted in 100 μL of 50% MeOH. Concatenate the 96 fractions into 24 samples by combining every other row [35], and concentrate again in vacuo to remove MeOH in the samples.
-
(f)
Determine the peptide concentration of each fraction using the BCA protein assay kit and dilute each of them to 0.1 μg/μL with H2O. Each fraction is analyzed using LC–MS/MS (see Subheadings 3.5 and 3.7).
-
(a)
3.3 Derivatization of Metabolites
-
1.
Make a methoxyamine solution with a pyridine concentration of 30 mg/mL.
-
2.
Dry aqueous metabolites briefly (30 min) in vacuo after removing from the freezer.
-
3.
Perform methoximation by adding 20 μL of methoxyamine solution to the sample vial and vortex for 30 s at setting 5 on a vortexer. Use a bath sonicator to ensure the sample is completely dissolved.
-
4.
Incubate the sample in a Thermomixer maintained at 37 °C for 1 h and 30 min with 1000 rpm shaking.
-
5.
Invert the vial one time to mix the samples with condensed drops at the cap surface.
-
6.
Spin the sample down for 1 min at 1000 × g at room temperature in a centrifuge.
-
7.
Perform silylation by adding 80 μL of N-methyl-N-trimethylsilyltrifluoroacetamide and 1% trimethylchlorosilane (MSTFA) solution using a syringe and vortex for 10 s.
-
8.
Incubate the sample in a Thermomixer maintained at 37 °C for 30 min with 1000 rpm shaking.
-
9.
Invert the vial one time to mix the samples with condensed drops at the cap surface.
-
10.
Spin the sample down for 5 min at 2000 × g at room temperature in a centrifuge.
-
11.
Transfer the reacted solution into a small volume insert and return to the same vial.
-
12.
Prepare a process blank sample using only reagents and FAMEs [27] as retention time alignment compounds (alternatively, a mixture of n-alkanes can be used) using the same protocol for the sample preparation.
-
13.
Tighten the caps and add to the autosampler tray for GC-MS analysis (see Subheading 3.4).
3.4 Metabolomics GC-MS Instrumental Analysis
-
1.
Install a nonpolar GC column in the GC oven (HP-5MS or DB-5MS, or similar polarity −30 m × 0.25 mm × 0.25 μm).
-
2.
Tune and calibrate the mass spectrometer (MS) before analysis to make sure the machine records the MS data correctly and check the helium gas pressure for possible leakage.
-
3.
Set up the optimized running parameters for the GC; injector: temperature at 250 °C, pressure and flowrate: 1 mL/min (or user defined value); oven temperature, ramping rate, holding time and the temperature of the MS transfer line should all be set at what was used to analyze the standard compounds used for constructing the reference database (if an in-house library is used).
-
4.
Set up the optimized running parameters in the MS; mass scan range (50–600 m/z), ion source temperature (250 °C), and ionization energy (70 eV) (the parameters which were used to run the standard compounds to build the database).
-
5.
Clean the injection syringe and plunger with acetone to remove any residue.
-
6.
Transfer the derivatized samples to the sample tray and place them in a randomized order to minimize instrumental artifacts.
-
7.
For large sample sets requiring more than 24 h to be analyzed, batching should be performed. Analyzing batches requires additional randomization and QC samples and/or internal standards to account for any type of variation that could be introduced through time. Necessary blanks and retention time standards need to be included in each batch being run on subsequent days.
3.5 Proteomics LC-MS/MS Instrumental Analysis
The LC-MS conditions presented in this section represent a typical method for analysis of isobaric labeled peptides (TMT-10plex™ Isobaric Labeling Reagent, Thermo Scientific). Use of this specific labeling reagent requires a MS capable of achieving >30k mass resolution in order to baseline resolve all reporter ions. However, other isobaric labeling reagents are available that do not require this level of resolution for labs not possessing instrumentation with this capability. Isobaric labeling strategies provide several benefits including multiplexing of large cohorts to improve throughput; use of a reference channel for data normalization; and improved quantitation due to elimination of run-to-run variation and reduction in sample processing variability. However, the data also suffer from “compression,” where signals from co-eluting peptides can affect reporter ion accuracy. There are several ways to mitigate this compression effect and one of the primary means is off-line fractionation of samples to reduce sample complexity. Another method uses the additional specificity of MS3 analysis (e.g., Thermo Scientific’s SPS MS3 workflow) where interference is reduced through a second MS/MS of primary fragment ions from the first MS/MS scan. This MS3 technique does not generally preclude the need for off-line fractionation. Interference can still occur, although it is generally minimized because the product ions selected for MS3 are more likely to be unique to a single peptide. The disadvantage of using MS3 is a significant hit on instrument duty cycle. It simply requires more time to perform the additional MS/MS event on primary fragment ions from every precursor ion. The method presented here uses only MS/MS, favoring an increased peptide identification rate over reduction in compression. For the purpose of this section, parameters for an M-Class nanoAcquity dual pumping UPLC (Waters, Milford, MA) coupled with a Q Exactive HF Hybrid Quadrupole-Orbitrap MS are described.
-
1.
UPLC Setup:
Prepare analytical and trapping columns in-house by slurry packing Jupiter 3 μm and 5 μm C-18 media into a 70-cm length of 360 μm o.d. × 75 μm i.d., and 4-cm length of 360 μm o.d. × 150 μm i.d. of fused silica, respectively. Use Kasil-based frits for retention of packing material.
-
(a)
Install the analytical and trap columns in a configuration such that once the sample has been trapped and washed, it is eluted in the opposite direction onto the analytical column.
-
(b)
Prepare mobile phases (MP); MP A: 0.1% formic acid in water and MP B: 0.1% formic acid in acetonitrile.
-
(c)
Set injection volume to 5 μL using a 5 μL sample loop and a 1.2× overfill value, resulting in a total of 7 μL of sample being pulled from the autosampler vial for each injection.
-
(d)
Set the trapping flow rate at 5 μL/min and a total trap time of 8 min.
-
(e)
Set the analytical flow rate to 300 nL/min with the following gradient and wash profile (min:%B); 0:1, 8:1, 10:8, 28:12, 83:30, 105:45, 108:95, 118:95, 122:50, 124:95, 126:1, 128:50, 130:50, 132:1, 152:1.
-
(f)
Events (min:event); 8:end trapping and start gradient, 28:start data acquisition, 108:end gradient and start column wash, 132:end column wash, 148:end data acquisition, 152:end column re-equilibration.
-
(a)
-
2.
Coupling UPLC to MS:
Couple the UPLC with the MS using a Nanospray Flex Ion Source, attaching the standard metal emitter tip that comes with the source to the end of the analytical column.
-
3.
MS Setup:
-
(a)
Perform required cleaning and/or calibrations. A full calibration is completed after the instrument has been deep cleaned (all optics cleaned up to and including the selection quadrupole). Otherwise, only mass calibrations are performed as needed (discussed in the QC section to follow).
-
(b)
Create a tune file that uses an ion transfer tube temperature of 325 °C and a spray voltage of 2 kV.
-
(c)
Create a data-dependent exclusion method with precursor settings using; mass range m/z 300–1800, resolution 60k, AGC target 3e6, max ion time 20 ms, and spectrum data type profile. For MS/MS settings, loop count 12, isolation width 0.7, resolution 30k, AGC target 1e5, max ion time 100 ms, fixed first mass m/z 110, normalized collision energy 30, and spectrum data type centroid. Additional settings include: min AGC target 5 × 103; intensity threshold 5 × 104; charge exclusion unassigned and 1; peptide match preferred; exclude isotopes on; dynamic exclusion 30 s; if idle do not pick others. Total acquisition time 120 min. Assume any other parameters listed were left with default settings.
-
(a)
-
4.
Quality Control (QC):
-
(a)
Add QC standard runs in a matter consistent with the goals of your research and the expected quality of your samples. If, during the course of method development, the samples are relatively “dirty,” then consider running QC standards daily or where reasonable between batches or blocks.
-
(b)
Select a QC standard that is similar to the samples that will be analyzed and assure enough material is on hand to conduct the entire study. For example, a 125 ng injection of RAW 264.7 macrophage cells (whole cell tryptic digest) would serve as a good QC standard for most mammalian cell studies (see Note 14).
-
(c)
Once a QC baseline has been established, run sample batches and blocks such that they are bracketed by QC standard runs. Only sample runs bracketed by acceptable QC standard runs are to be used (see Note 15).
-
(d)
Visually inspect each run, looking for shifts in retention time of common peaks and changes in signal stability and intensity.
-
(e)
Monitor mass accuracy by calculating the mass error for a commonly observed contaminant (e.g., polysiloxane) that produces a singly charged ion having a mass (m/z) of 445.1200.
-
(f)
QC samples should be run at least weekly, between sample batches, or anytime instrument performance is in question.
-
(a)
3.6 Lipidomics LC-MS/MS Instrumental Analysis
-
1.
The specific conditions listed below are for a Waters Acquity UPLC H class system and Velos Pro Orbitrap mass spectrometer (see Note 16).
-
2.
Set up and purge the LC system with lipid mobile phase A and B for 2 min or until all lines within the flow path have been cleared.
-
3.
Install an analytical LC column (Waters CSH 3.0 mm × 150 mm, 1.7 μm particle size) and set the column temperature at 42 °C and the injection volume to 10 μL.
-
4.
Set the flow rate to 250 μL/min and build the gradient profile as follows: (min, %B): 0, 40; 2, 50; 3, 60; 12, 70; 15, 75; 17, 78; 19, 85; 22, 92 25, 99; 34, 99; 34.5, 40.
-
5.
Tune and calibrate the mass spectrometer according to the manufacturer’s recommendations.
-
6.
Set up the source parameters as follows: MS inlet and source 350 °C, spray voltage of 3.5 kV, sheath gas flow of 45, auxiliary gas flow of 30, and sweeping gas flow of 2.
-
7.
Analyze each sample in both positive and negative ion mode with a precursor scan range of 200–2000 m/z and at a mass resolution of 60k.
-
8.
Set up a data-dependent MS/MS method consisting of fragmentation of the top four ions, alternating between HCD and CID modes, and with the following dynamic exclusion parameters: repeat count 1, repeat duration 15 (s), exclusion list size of 250, exclusion duration of 8 (s), a low exclusion mass width of 0.55 and a high exclusion mass width of 1.5.
-
9.
Acquire CID spectra using a minimum signal threshold of 500, isolation width of 2 m/z, normalized collision energy of 35, default charge state of 2 (see Note 17), activation Q value of 0.18 and activation time of 10.0 (ms).
-
10.
Acquire HCD spectra at a resolution of 7500, a minimum signal threshold of 1000, isolation width of 2 m/z, normalized collision energy of 30, default charge state of 2, activation time of 0.100 (ms), and the first mass fixed at 90 m/z.
-
11.
Couple the LC to the mass spectrometer and acquire three QC samples as a means of checking LC and instrument performance. The BTLE QC sample is injected and analyzed together with the samples as a means to monitor system performance, such as retention time, peak intensity, peak shape, and mass measurement error using data acquisition software. Furthermore, the BTLE QC sample is injected and analyzed after cleaning and calibration of the instrument, replacing the analytical column or any maintenance performed on the system.
-
12.
Add the following sequence to the beginning and end of the sample queue to monitor carryover and instrument performance: methanol blank, QC, methanol blank.
-
13.
For large sample sets insert the sequence from step 12 throughout the queue as necessary.
-
14.
Total Lipid Extracts (TLEs) are stored in 2:1 chloroform:methanol and evaporated in vacuo, then reconstituted in 5 μL chloroform and 45 μL of methanol prior to injecting 10 μL onto the column.
3.7 Protein Identifications
For the proteomics data analysis, there is a variety of commercial and open-source tools currently available. The analysis parameters are also by the preference of the user. In our laboratory, we regularly perform proteomic analysis with a combination of msConvert (for spectrum to peak list conversion) [28], mzRefinery (for mass recalibration) [29], MS-GF+ (for peptide/protein identification) [30], and MASIC (for quantification) [31].
-
1.
Peak list conversion with msConvert parameters:
-
(a)
Convert to _DTA.txt format.
-
(b)
Convert to centroid peak.
-
(a)
-
2.
Mass recalibration with mzRefinery parameters:
-
(a)
Same searching parameters described below.
-
(a)
-
3.
Peptide/protein identification with MS-GF+:
-
(a)
Protein sequence database. The database can be downloaded from a variety of sources, such as RefSeq, UniprotKB, and Ensembl. For human cells, we recommend using Swiss-Prot, which is available at www.uniprot.org.
-
(b)
Searching parameters:
-
Enzyme: Trypsin.
-
Number of tolerable termini: 1 (partial trypsin digestion).
-
Minimum peptide length: 6.
-
Maximum Peptide Length: 50.
-
Number of matches per spectrum: 1.
-
-
Variable modifications: methionine oxidation (+15.9949 Da).
-
Fixed modifications: cysteine carbamidomethylation (+57.0215).
-
Parent mass tolerance: 20 ppm.
-
Isotope error: −1 and 1.
-
Targeted Decoy Analysis: true.
-
Fragmentation method: automatic detected (as written in the spectrum).
-
Instrument: QExactive.
-
Filter searching results based on MS-GF probability score. Usually, a score ≤1 × 10−10 results in a false-discovery rate ≤1%.
-
Extraction of quantitative information with MASIC. Parameters:
-
Mass tolerance for reporter ions: 0.003 m/z.
-
The identified peptides and reporter ion intensities can be combined using Microsoft Access. Sum the intensities of multiple spectra of the same peptide and multiple peptides of the same protein.
-
-
Expected results: the number of identified peptides and proteins will vary according to the mass spectrometer and peptide separation strategy. For instance, mammalian cells infected with MERS-CoV prefractionated into 24 fractions and analyzed in a 100-min gradient on a QExactive mass spectrometer should lead to the identification of 8000–10,000 proteins.
-
Troubleshooting: If the number of identified proteins is much smaller than expected, check the following items:
-
Contamination: detergents and plasticizers are common contaminants in samples and they have a characteristic profile in LC-MS/MS data by having uniform mass and retention time shifts. The solution to this problem is to eliminate the source of these contaminants by washing the glassware, avoiding detergents during sample preparation steps and checking the compatibility of used plastic tubes with the extraction solvents.
-
Searching parameters: another source of poor coverage can be due to incorrect searching parameters. Check the sequence database (if correct species was used, for instance), enzyme specificity, and peptide modifications.
-
-
-
(a)
3.8 Metabolite Identifications
-
1.
Check that all of the data files were correctly obtained from the analysis. If an internal standard(s) was spiked, make sure the retention time and peak intensity values are consistent throughout the sample analysis.
-
2.
Convert the MS raw data into a general MS format if required (e.g., netCDF).
-
3.
Upload all the data files into the chosen software (e.g., Metabolite Detector, MassHunter, AMDIS, or similar) and perform an alignment of retention time using the acquired data from the analysis of retention time alignment compounds (e.g., FAMEs or Alkane mix).
-
4.
Align the retention time information to all the collected data files, and perform library matching in order to identify metabolites.
-
5.
Perform a batch analysis to align all the detected metabolite peaks.
-
6.
Export the processed data into a CSV format, and read the data from a spreadsheet program (e.g., Excel).
-
7.
Check the matching scores of identified metabolites and their signal-to-noise ratios (higher than 75/100 matching score, with S/N > 3). Select the correctly identified metabolites for further biological interpretation (e.g., integrated analysis with other omics data including proteomics and lipidomics).
3.9 Lipid Identifications
-
1.
Launch LIQUID program [33] and upload a .raw or .mzXL data file using the drop down menu.
-
2.
Select the ionization mode the samples were acquired in and enter mass error tolerances based on instrument QCs and sample data.
-
3.
Load lipid targets, set results per scan to 1, then process targets.
-
4.
Sort the list of potential lipid identifications by common name, and then scan number.
-
5.
Inspect the potential identifications one at a time, examining and evaluating the following:
-
(a)
Diagnostic ion and associated fragment ions (e.g., fatty acids). Verify that the expected peaks are present.
-
(b)
Isotopic profile: it should match predicted profile.
-
(c)
Extracted ion chromatogram: confirm the parent scan is at the peak apex.
-
(d)
Mass error: confirm it is consistent with the rest of the data.
-
(e)
Retention time: verify that it is within allowable tolerances for LC system.
-
(a)
-
6.
Select the correct identifications as going through step 5.
-
7.
Export selected results.
-
8.
Using LIQIUD’s exported results from step 7, create a target list including observed m/z, retention time, and common name.
-
9.
Launch MZmine and import data files.
-
10.
Associate targeted files from step 8 to the raw data files imported in step 9.
-
11.
Perform mass detection on files not analyzed in LIQUID, and then build chromatograms from generated mass list.
-
12.
Perform peak alignment and then gap-filling on the aligned list.
-
13.
Check feature alignment and manually verify correct alignment for each lipid identification.
-
14.
Export final results including lipid name and peak area or peak apex intensity, which can be used for statistical analysis.
4 Notes
-
1.
18.2 MΩ cm water (which will be referred to as water) should be used to prepare all solutions unless otherwise noted.
-
2.
The chloroform:methanol solution should always be at least −20 °C to facilitate protein precipitation.
-
3.
Order the highest purity possible.
-
4.
Purchase MSTFA with 1% TMCS in a 5 mL size. Once it is opened, moisture in the air can reduce the reactivity of the reagent. It can cause skin corrosion, serious eye damage, and specific target organ toxicity, and is a flammable liquid and vapor. Wear safety glasses, gloves, and lab coat, and work in a fume hood.
-
5.
All tryptic digestion chemicals are purchased from Sigma-Aldrich unless otherwise listed.
-
6.
Lysine residues modified by reductive methylation, yielding a highly active and stable trypsin.
-
7.
Polypropylene is conditionally resistant to chloroform and can be used as long as contact time is minimized. A fresh tip must be used each time after chloroform is dispensed.
-
8.
The Sorenson brand tubes have shown to be compatible with chloroform and do not leach polymers that can interfere with downstream analysis.
-
9.
Three sets of Sorenson tubes should be generated for each sample. Samples can be collected and MPLExed in the tubes labeled for protein/proteomics as the protein remains in the tube following removal of the aqueous (into a tube labeled metabolites) and organic (into a tube labeled lipids) phases (see Fig. 1).
-
10.
Aliquots of samples are removed and tested at this step to ensure all virus has been inactivated.
-
11.
At this point the sample is inactivated and separated into three distinct analytes and each of them can be handled in BSL1 conditions.
-
12.
BCA assay regent is compatible up to 3 M urea, so when the sample is in 8 M urea, appropriate dilution factors should be considered.
-
13.
The TMT Reagents are amine-reactive and modify lysine residues and the peptide N-termini. All amine-containing buffers and additives must be removed before digestion and labeling.
-
14.
Prior to beginning a study, determine acceptable values for a selected set of metrics for the QC standard to be used. This should be performed using a clean instrument and new analytical and trap columns. Example metrics to monitor for performance are number of unique peptides identified, average chromatographic peak width, and mass error. QC procedures for complex proteomics measurements are not always straightforward. Establish a baseline for the QC process before running samples.
-
15.
Always run a blank before and after QC standard runs.
-
16.
In general, an LC capable of delivering a flow rate of 250 μL/min with a pressure limit of 8000 psi or greater and a mass spectrometer with a mass resolution of 30k or higher is required for this method.
-
17.
The default charge state of 2 is used instead of 1 to ensure we don’t miss the doubly charged lipids (e.g., cardiolipins and gangliosides) and the detection of single charged lipids by setting the default charge state to 2 is unaffected.
References
Modjarrad K et al (2016) A roadmap for MERS-CoV research and product development: report from a World Health Organization consultation. Nat Med 22(7):701–705
Aderem A et al (2011) A systems biology approach to infectious disease research: innovating the pathogen-host research paradigm. MBio 2(1):e00325–e00310
Cockrell AS et al (2016) A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol 2:16226
Menachery VD et al (2018) MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc Natl Acad Sci U S A 115(5):E1012–E1021
Nakayasu ES et al (2016) MPLEx: a Robust and universal protocol for single-sample integrative proteomic, metabolomic, and lipidomic analyses. mSystems 1(3):e00043
Burnum-Johnson KE et al (2017) MPLEx: a method for simultaneous pathogen inactivation and extraction of samples for multi-omics profiling. Analyst 142(3):442–448
Menachery VD et al (2014) Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. MBio 5(3):e01174–e01114
Mitchell HD et al (2013) A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS One 8(7):e69374
Gralinski LE et al (2013) Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. MBio 4(4):e00271
Sims AC et al (2013) Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J Virol 87(7):3885–3902
Kyle JE et al (2019) Plasma lipidome reveals critical illness and recovery from human Ebola virus disease. Proc Natl Acad Sci U S A 116(9):3919–3928
Eisfeld AJ et al (2017) Multi-platform omics analysis of human ebola virus disease pathogenesis. Cell Host Microbe 22(6):817–829 e8
Tisoncik-Go J et al (2016) Integrated omics analysis of pathogenic host responses during pandemic H1N1 influenza virus infection: the crucial role of lipid metabolism. Cell Host Microbe 19(2):254–266
Nicod C, Banaei-Esfahani A, Collins BC (2017) Elucidation of host-pathogen protein-protein interactions to uncover mechanisms of host cell rewiring. Curr Opin Microbiol 39:7–15
Wang YP, Lei QY (2018) Metabolite sensing and signaling in cell metabolism. Signal Transduct Target Ther 3:30
DeBerardinis RJ, Thompson CB (2012) Cellular metabolism and disease: what do metabolic outliers teach us? Cell 148(6):1132–1144
Sanchez EL, Lagunoff M (2015) Viral activation of cellular metabolism. Virology 479:609–618
Wenk MR (2006) Lipidomics of host-pathogen interactions. Chem Phys Lipids 143(1-2):42–42
Knoops K et al (2008) SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6(9):1957–1974
Knoops K et al (2010) Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J Virol 84(2):833–846
Ulasli M et al (2010) Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol 12(6):844–861
de Wilde AH et al (2013) MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol 94:1749–1760
Miller S, Krijnse-Locker J (2008) Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6(5):363–374
Stapleford KA, Miller DJ (2010) Role of cellular lipids in positive-sense RNA virus replication complex assembly and function. Viruses 2(5):1055–1068
Blanchard E, Roingeard P (2015) Virus-induced double-membrane vesicles. Cell Microbiol 17(1):45–50
Folch J, Lees M, Stanley GHS (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226(1):497–509
Kind T et al (2009) FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem 81(24):10038–10048
Adusumilli R, Mallick P (2017) Data conversion with ProteoWizard msConvert. Methods Mol Biol 1550:339–368
Gibbons BC et al (2015) Correcting systematic bias and instrument measurement drift with mzRefinery. Bioinformatics 31(23):3838–3840
Kim S, Pevzner PA (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun 5:5277
Monroe ME et al (2008) MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput Biol Chem 32(3):215–217
Hiller K et al (2009) MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem 81(9):3429–3439
Kyle JE et al (2017) LIQUID: an-open source software for identifying lipids in LC-MS/MS-based lipidomics data. Bioinformatics 33(11):1744–1746
Pluskal T et al (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395
Wang Y et al (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11(10):2019–2026
Acknowledgments
The methods described in this chapter were supported in part by National Institute of Allergy and Infectious Diseases grant U19AI106772. Additional support was provided by National Institute of General Medical Sciences Grant P41 GM103493. Lipidomics, proteomics, and metabolomics analyses were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy (DOE) Office of Biological and Environmental Research, and located at Pacific Northwest National Laboratory (PNNL). PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Nicora, C.D. et al. (2020). Metabolite, Protein, and Lipid Extraction (MPLEx): A Method that Simultaneously Inactivates Middle East Respiratory Syndrome Coronavirus and Allows Analysis of Multiple Host Cell Components Following Infection. In: Vijay, R. (eds) MERS Coronavirus. Methods in Molecular Biology, vol 2099. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0211-9_14
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0211-9_14
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0210-2
Online ISBN: 978-1-0716-0211-9
eBook Packages: Springer Protocols