Abstract
Three-dimensional (3D) reconstruction is an essential approach in morphological studies in biology and paleontology. Seeking an optimized protocol for nondestructive observations, we attempted 3D visualization of various molluscan shells and animals with X-ray micro-computed tomography (micro-CT). Calcified parts of molluscs were easily visualized except for cases with marked differences in thickness heterogeneity. 3D imaging of shell microstructure was difficult. Visualization of soft tissue requires staining to enhance the image contrast. Especially for soft tissues, synchrotron X-ray microtomography is the most advanced method to generate clear 3D images. 3D data facilitates morphological quantification, enabling calculations of length and volume even for very complex forms. X-ray micro-CT is extremely useful in the morphologic examination of mineralized and soft tissues, although microstructural and histological details should be supplemented by other microscopic techniques.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The importance of computed tomography (CT) has been established in biology and paleontology. CT has also been used in examinations of vertebrate morphology (Gignac et al. 2016; du Plessis et al. 2017), and micro-CT has been used in invertebrate biology, mainly in entomology studies (Friedrich et al. 2014; Wipfler et al. 2016). The approach has been infrequently used for other animal groups. In malacology, the use of micro-CT is restricted to a small number of case studies involving anatomy (Golding and Jones 2007; Golding et al. 2009), ontogeny (Kerbl et al. 2013), paleontology (Takeda et al. 2016), and shell morphology (Monnet et al. 2009; Liew and Schilthuizen 2016; Noshita et al. 2016).
The application of CT is still in the testing and development stage. Having an optimized protocol for the nondestructive micro-CT analysis of various morphological characteristics would help expand the scope of CT applications and was the focus of the present study.
2 Material and Methods
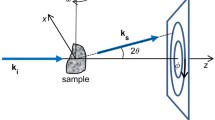
The molluscan specimens used in this study are all registered and deposited in The University Museum, The University of Tokyo (UMUT). The abbreviations RM and CM in the registration numbers indicate recent and Cenozoic molluscs, respectively. Animals were fixed in 5% formaldehyde solution for several days, washed in tap water, and then preserved in 70% ethanol. Intact animals were extracted from shells by soaking the specimens in approximately 10% HCl to completely dissolve the shells. Most specimens were first digitally imaged (Fig. 9.1a–e) using a ScanXmate B100TSS110 industrial micro-CT device (Comscantecno Co., Ltd.) at UMUT. Scan parameters were adjusted according to samples and included source voltage (70–100 kV), source current (approximately 40–150 μA), and exposure time per frame (0.4–1.0 s). The total number of frames was 1200, the number of pixels of the detector was 1024 × 1012, and the highest resolution was approximately 2 μm. Samples larger than 3 cm were scanned in the laboratory of Comscantecno Co. Ltd. For scanning, embedding the sample in a heat-generated hole in Styrofoam on a rotating stage was the most effective method of sample fixation. For soft portions of samples, high-resolution X-ray CT was also performed (Fig. 9.1f–h) at the hutch 3 of the beamline BL20B2 in SPring-8 (Hyogo, Japan) with a resolution of 13.16 μm per pixel and an X-ray energy of 25 keV. Three-dimensional (3D) visualization was conducted using Molcer Plus (White Rabbit Corporation), OsiriX (OsiriX Foundation), and Amira 3.5.5 (Visage Imaging, Inc.) software.
3 Results
3.1 Shells
Most samples of calcified shells were scanned without difficulty using conventional industrial micro-CT (Fig. 9.2). The internal structure of shells was also perfectly reconstructed from scan data. Exceptions included shells of specific groups of gastropods, with shells from different regions having extremely disparate thicknesses. For example, members of the families Ellobiidae (Figs. 9.2c, d), Olividae, and Conidae have shells in which the internal whorls are much thinner than the outer wall owing to the secondary resorption that occurs during growth. In such cases, the thinner inner whorls disappear or became patchy in distribution when the settings were optimized for the outer shell surface. Conversely, if contrast were enhanced to reveal the inner structures, outer surfaces were rendered unusable by the increased noise levels (the overexposure effect). So far, no viable solution to this problem has been identified in our current system.
Example of 3D reconstruction of calcified shells from industrial micro-CT data. (a, b) Volutoconus bednalli (Brazier, 1878) (Volutidae). UMUT RM32762. (c, d) Ellobium aurismidae (Linnaeus, 1758) (Ellobiidae). UMUT RM32763. Arrowhead indicates missing inner wall as artifact. (e, f) Cerithiella trisulcata (Yokoyama, 1922) (Newtoniellidae). UMUT CM20781. Holotype. (g–i) Tiberia pseudopulchella (Yokoyama, 1920) (Pyramidellidae). UMUT CM20242. Holotype. (i) Apical view. (j) Adapical view. Shell height: a, b = 90.6 mm; c, d 88.8 mm; e, f = 5.2 mm; g, h = 7.0 mm. Software: Molcer Plus
Digitization using micro-CT is an excellent approach to display the overall 3D morphology and the internal structure of molluscan shells. Examples of specimens of small Cenozoic fossil gastropods are presented in Fig. 9.2e–j. Virtual slicing of type specimens along an arbitrary plane is possible only using micro-CT. Prominent columellar folds (Fig. 9.2f, h) or denticles in the outer lip (Fig. 9.2h) were important features to morphologically diagnose species. Construction of images from apical (Fig. 9.2i) and basal (Fig. 9.2j) views for fragile and minute specimens is easily acquired. However, imaging of the same specimens by photography with a digital camera and binocular microscope carries the risk of specimen loss or damage. Micro-CT allows the detailed nondestructive observation of the delicate structures of shells.
However, the use of micro-CT for shells is not entirely versatile. Most importantly, growth lines, shell layers, and shell microstructure cannot be observed. We were unable to detect any structure on sliced shell planes in the CT-derived 3D data. The observation of these fine structures still necessitates mechanical destruction or cutting of the actual shell samples.
Another issue is that high-resolution scanning of a specific part of a large shell is not possible. To achieve high-resolution scan data, it is necessary to place a sample as close to the X-ray source as possible (for any given detector image size, the realized resolution depends on the field of view and, thus, the size of the specimen). In conventional micro-CT, it is necessary to rotate the specimen. Thus, if the sample is large, it must be placed further away from the X-ray source, which will decrease the resolution. This dilemma cannot be overcome at the present time.
3.2 Soft Tissues
Simultaneous visualization of both the shell and the soft regions from a single specimen could not be achieved with satisfactory results. If various parameters are optimized for shells, the settings are far from optimal for the soft regions, and vice versa. The only way to achieve high-quality scans for both features from a single individual is to obtain the shell scan first and to decalcify it to scan the soft regions. This destroys the shell. Therefore, development of a new algorithm or methodology is desired to enable the nondestructive scans of precious specimens (such as intact types) to reveal the anatomy of soft portions.
Applying CT to soft parts is more challenging than applying it to calcified parts. Animal tissues usually do not display any contrast in CT images without the aid of contrast-enhancing substances. Figure 9.3 shows an example of Patellogastropoda. In this organism, only iron-mineralized radular teeth are visible if the specimen is not treated with a contrast agent (e.g., iodine).
Installation of samples on the rotating stage is also an extremely sensitive part of the scanning process. If a wet sample is scanned while it is exposed to air, it quickly becomes dehydrated and deformed, which will result in a very blurred image that is unusable. Therefore, samples should be contained in a liquid medium such as ethanol, water, or physiological saline water in a (relatively) X-ray transparent container. Wrapping a sample with X-ray transparent film in a Styrofoam container can be an effective solution. Expelling air bubbles from the medium is essential because the bubbles can cause sample movement when they expand as they are subjected to heat generated during prolonged scanning.
In our experience, despite repeated experiments, scanning of soft parts with a conventional industrial micro-CT frequently resulted in insufficient contrast and clarity (Fig. 9.4). To explore other solutions, we attempted to use synchrotron CT instead of industrial CT (Fig. 9.1f–h). The results were clearly superior to the industrial CT data, as exemplified in an experiment in which the same specimen was used for both methods (Fig. 9.5). The synchrotron CT data were characterized by the clear edge of the outline and sharp boundaries of the internal organs. The configurations of internal organs were more reliably identified using this method.
Soft part of Lottia dorsuosa (Gould, 1859) (Lottiidae). UMUT RM32764. Comparison of scanning results between conventional industrial micro-CT (a) and synchrotron CT (b SPring-8 BL20B2). dg digestive gland, es esophagus, f foot, i intestine, mm mantle margin, pb pedal blood sinus, pc pallial cavity, rds radular sac, st stomach. Body length = 22 mm
A widely utilized method that was also used by us is to immerse samples in 1% iodine solution as the contrast agent for both laboratory and synchrotron CT scanning. This staining method is very handy and inexpensive. In our experiments, iodine rapidly penetrated into the samples, and immersion in iodine for 1 to several days was sufficient for samples smaller than 2 cm. However, slight shrinkage of the samples was unavoidable with iodine staining. This artifact was especially evident with the thin and membranous mantle tissue, which displayed numerous minute wrinkles after staining with iodine. In this study, we could not sufficiently test different contrast agents such as phosphomolybdic acid and osmium tetroxide. This will be the subject of a future study.
We tested Molcer Plus, OsiriX, and Amira software. Each has its own inherent advantages. Molcer Plus was the best software for visualizing calcified shells. OsiriX was able to create vivid 3D images of soft portions (Fig. 9.6b–d) with the most straightforward operation. Amira was superior in terms of results and operation for internal anatomy (Fig. 9.6e, f). By adjusting the threshold of brightness, the surface outline of a specimen could be automatically extracted (Figs. 9.6e and 9.7a–c). Different parts of internal organs could be illustrated in various colors after the segmentation procedure (Fig. 9.6f). Figure 9.7 is an example of the 3D visualization from three different angles, showing the animal’s surface outline, digestive tracts with a translucent outline, and isolated digestive tracts only. This method allowed us to quantify the size and volume of various parts of the animal body.
Examples of 3D visualization of soft parts from synchrotron CT data. (a–d) Spirostoma japonicum (A. Adams, 1867) (Cyclophoridae). UMUT RM32766. Body length = 9.7 mm. (a) Transmission image. (b) Ventral view, (c) dorsal view, (d) frontal view. (e, f) Granata lyrata (Pilsbry, 1890) (Chilodontidae). UMUT RM32767. Body length = 9.9 mm. (e) Body surface. (f) Digestive tracts with translucent body surface. Software: (a–d) OsiriX. (e, f) Amira
Examples of 3D visualization of soft parts from synchrotron CT data (SPring-8 BL20B2). Phenacolepas unguiformis (Gould, 1859) (Phenacolepadidae). UMUT RM32768. (a–c) Exterior. (d–f) Digestive tracts with translucent outline. (g–i) Extracted digestive tracts. (a, d, g) Dorsal view. (b, e, h) Left lateral view. (c, f, i) Ventral view. Esophagus (blue), stomach (red), and intestine (yellow) are indicated in different colors. Body length = 6.0 mm. Software: Amira
4 Discussion
Through this experimental study, we confirmed that micro-CT is a very useful method for 3D presentation of various hard and soft features of molluscs. CT scanning has three important advantages. First, observations of internal shell structures are possible without having to cut the shells. Second, acquisition of a complete series of serially sectioned images without damaging soft portions is feasible. Third, the use of synchrotron CT allows 3D reconstruction of soft parts in much less time.
Scanning calcium carbonate shells is relatively easily performed and should be applied widely to other biominerals of invertebrates such as corals, barnacles, and echinoderms (Okanishi et al. 2017). The most comprehensive method for understanding shell morphology is achieved by the combination of micro-CT and scanning electron microscopy (SEM). Micro-CT has the advantages of 3D digitization and nondestructive internal observation. For example, it is possible to reveal internal morphological characteristics of specimens without cutting through them (Fig. 9.2) and can be achieved from any direction and for specimens of any size. On the other hand, SEM has a much higher resolution, which enables the observation of microscopic sculptures at higher magnification. Therefore, SEM is still necessary for characterizing micromolluscs (Sasaki 2008). In addition, shell microstructure and shell layer structure are most reliably observed with SEM (Nishida et al. 2012; Sato and Sasaki 2015).
We confirmed that micro-CT is also extremely effective for the study of soft parts. In particular, this method has a tremendous benefit for samples that are difficult to section with a microtome. For example, the digestive tracts of deposit feeders such as infaunal bivalves are often filled with many sediment grains, which easily damage knives during histological sectioning. In patellogastropod limpets (Fig. 9.3) and chitons, the radular teeth are heavily mineralized and also cause serious knife damage. Using micro-CT avoids these problems.
Before the introduction of micro-CT, serial histological sectioning was the only method available for 3D reconstruction (Chen et al. 2015; see Ruthensteiner 2008 for detailed methodology). However, with micro-CT, we are liberated from the difficult and time-consuming demand of making a complete series of serial sections of whole animals. For 3D reconstruction from serial sections, considerable time is spent aligning the stacks. This issue does not exist in micro-CT because the image stack generated is completely aligned without skew. In addition, complete anatomical reconstruction of large specimens becomes possible (the realistic upper size limit of resin-embedded serial sectioning is approximately 5 mm wide, and paraffin sections are too skewed to be useful for 3D visualization). Therefore, CT presents a high-throughput, highly efficient approach compared to sectioning, as has previously been pointed out by Golding and Jones (2007) and Kerbl et al. (2013).
Generating 3D data from a 2D image stack is still a time-consuming process with regard to internal anatomy. At present, we do not have a practical algorithm for automatic segmentation of the various internal organs. This continues to hinder advances in 3D morphological analysis.
The resolution of CT data is still much lower than that of histological sectioning. In addition, we cannot distinguish different organs using distinctive staining, as is used for paraffin-embedded sectioning or immunohistochemistry. For example, muscle and connective tissue can be vividly distinguished by trichrome staining (Katsuno and Sasaki 2008), but such a distinction is not possible with CT data. Therefore, it is advisable to reconstruct various 3D morphologies from CT data and supplement the details from histological sections. In this approach, much less histology data are needed for an overall understanding of the specimen. In this way, we can greatly improve the efficiency of morphological studies with CT.
3D data with segmentation may be exploited for various morphological analyses, especially quantitative analysis. For example, we can calculate the length and volume of various structures that are difficult to measure in actual specimens. Quantification of shape and its applications are topics for the future.
In conclusion, X-ray micro-CT is extremely useful in the morphological characterization of both mineralized and non-mineralized tissues of molluscs. Although this method has the potential to greatly improve the efficiency of morphological studies, it does not entirely replace conventional approaches and should be supplemented by data from other destructive microscopic methods.
References
Chen C, Copley JT, Linse K, Rogers AD, Sigwart JD (2015) The heart of a dragon: 3D anatomical reconstruction of the ‘scaly-foot gastropod’ (Mollusca: Gastropoda: Neomphalina) reveals its extraordinary circulatory system. Front Zool 12(13):1–16
du Plessis A, Broeckhoven C, Guelpa A, le Roux SG (2017) Laboratory x-ray micro-computed tomography: a user guideline for biological samples. Gigascience 6:1–11
Friedrich F, Matsumura Y, Pohl H, Bai M, Hoernschemeyer T, Beutel RG (2014) Insect morphology in the age of phylogenomics: innovative techniques and its future role in systematics. Entomol Sci 17:1–24
Gignac PM, Kley NJ, Clarke JA, Colbert MW, Morhardt AC, Cerio D, Cost IN, Cox PG, Daza JD, Early CM, Echols MS, Henkelman RM, Herdina AN, Holliday CM, Li Z, Mahlow K, Merchant S, Mueller J, Orsbon CP, Paluh DJ, Thies ML, Tsai HP, Witmer LM (2016) Diffusible iodine-based contrast-enhanced computed tomography (diceCT): an emerging tool for rapid, high-resolution, 3-D imaging of metazoan soft tissues. J Anat 228:889–909
Golding RE, Jones AS (2007) Micro-CT as a novel technique for 3D reconstruction of molluscan anatomy. Molluscan Res 27:123–128
Golding RE, Ponder WF, Byrne M (2009) Three-dimensional reconstruction of the odontophoral cartilages of Caenogastropoda (Mollusca: Gastropoda) using micro-CT: morphology and phylogenetic significance. J Morphol 270:558–587
Katsuno S, Sasaki T (2008) Comparative histology of radula-supporting structures in Gastropoda. Malacologia 50:13–56
Kerbl A, Handschuh S, Noedl M-T, Metscher B, Walzl M, Wanninger A (2013) Micro-CT in cephalopod research: investigating the internal anatomy of a sepiolid squid using a non-destructive technique with special focus on the ganglionic system. J Exp Mar Biol Ecol 447:140–148
Liew TS, Schilthuizen M (2016) A method for quantifying, visualising, and analysing gastropod shell form. PLoS One 11:e0157069
Monnet C, Zollikofer C, Bucher H, Goudemand N (2009) Three-dimensional morphometric ontogeny of mollusc shells by micro-computed tomography and geometric analysis. Palaeontol Electron 12.3(12A):1–13
Nishida K, Ishimura T, Suzuki A, Sasaki T (2012) Seasonal changes in the shell microstructures of the bloody clam, Scapharca broughtonii (Mollusca: Bivalvia: Arcidae). Palaeogeogr Palaeoclimatol Palaeoecol 363–364:99–108
Noshita K, Shimizu K, Sasaki T (2016) Geometric analysis and estimation of the growth rate gradient on gastropod shells. J Theor Biol 389:11–19
Okanishi M, Fujita T, Maekawa Y, Sasaki T (2017) Non-destructive morphological observations of the fleshy brittle star, Asteronyx loveni using micro-computed tomography (Echinodermata, Ophiuroidea, Euryalida). ZooKeys 663:1–19
Ruthensteiner B (2008) Soft part 3D visualization by serial sectioning and computer reconstruction. Zoosymposia 1:63–100
Sasaki T (2008) Micromolluscs in Japan: taxonomic composition, habitats, and future topics. In: Geiger DL, Ruthensteiner B (eds) Micromollsucs: methodological challenges – exciting results. Zoosymposia 1:147–232
Sato K, Sasaki T (2015) Shell microstructure of Protobranchia (Mollusa: Bivalvia): diversity, new microstructures and systematic implications. Malacologia 59(1):45–103
Takeda Y, Tanabe K, Sasaki T, Uesugi K, Hoshino M (2016) Non-destructive analysis of in situ ammonoid jaws by synchrotron radiation X-ray micro-computed tomography. Palaeontol Electron 19.3(46A):1–13
Wipfler B, Pohl H, Yavorskaya MI, Beutel RG (2016) A review of methods for analysing insect structures – the role of morphology in the age of phylogenomics. Curr Opin Insect Sci 18:60–68
Acknowledgments
Dr. Osamu Sasaki (The Tohoku University Museum) and Dr. Akiteru Maeno (National Institute of Genetics) kindly provided technical advice when we introduced the Comscantechno CT scanner to The University Museum, The University of Tokyo. This study was funded by a JSPS Kakenhi Grant, number 15K14589 and JP16J06269. The synchrotron radiation experiments were performed at the BL20B2 of SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal Nos. 2015B1833, 2016A1706, 2017A1720, 2017B1767).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2018 The Author(s)
About this paper
Cite this paper
Sasaki, T. et al. (2018). 3D Visualization of Calcified and Non-calcified Molluscan Tissues Using Computed Tomography. In: Endo, K., Kogure, T., Nagasawa, H. (eds) Biomineralization. Springer, Singapore. https://doi.org/10.1007/978-981-13-1002-7_9
Download citation
DOI: https://doi.org/10.1007/978-981-13-1002-7_9
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1001-0
Online ISBN: 978-981-13-1002-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)