Abstract
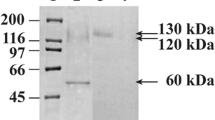
The glucoamylase gene of the yeast Arxula adeninivorans Ls3 has been cloned from a genomic library and sequenced. The gene could be localized on chromosome 2 from A. adeninivorans and comprises 1875 bp. The first 16 N-terminal amino acids represent the signal sequence for entering the endomembrane system. Comparing the amino acid sequence from this glucoamylase with those of other fungal glucoamylases shows that the glucoamylase of strain Ls3 has a homology to the glucoamylases from Rhizopus oryzae (32.6%), Saccharomycopsis fibuligera (23.1%), Aspergillus niger (22.1%), and Saccharomyces diastaticus (15.4%). No homology could be detected to the glucoamylase of Schwanniomyces occidentalis. By using the GAL1 promoter from Saccharomyces cerevisiae within an autonomously replicating plasmid it was possible to express the isolated Arxula glucoamylase gene in Saccharomyces cerevisiae. The transformants secreted 95% of the enzyme into the culture medium. The N termini of glucoamylases synthesized in A. adeninivorans and S. cerevisiae transformants are identical, which means that the signal sequences were cleaved at the same positions during maturation of the proteins. The highest glucoamylase activities were reached in the culture medium of S. cerevisiae transformants after 36 h of fermentation. Northern hybridization showed that the glucoamylase transcripts were formed continuously for up to 70 h. These results reveal that the glucoamylase is expressed and secreted more rapidly in the S. cerevisiae transformants than in A. adeninivorans Ls3.
Similar content being viewed by others
References
Anbertin AM, Tondre C, Lopez C, Obert G, Kirn A (1983) Sodium dodecyl sulfate-mediated transfer of electrophoretically separated DNA binding proteins. Anal Biochem 131:127–134
Ashikari T, Nakamura N, Tanaka Y, Kiuchi N (1986) Rhizopus raw-starch-degradation glucoamylase—its cloning and expression in yeast. Agric Biol Chem 50:957–964
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523
Boel H, Hjort I, Svensson B, Noriss F, Noriss KE, Fiil NB (1984) Glucoamylases G1 and G2 from Aspergillus niger are synthesized from two different but closely related mRNA. EMBO J 3:1097–1102
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Büttner R, Bode R, Birnbaum D (1987) Purification and characterization of the extracellular glucoamylase from the yeast Trichosporon adeninovorans. J Basic Microbiol 27:299–308
Büttner R, Bode R, Birnbaum D (1991) Comparative study of external and internal β-glucosidases and glucoamylase of Arxula adeninivorans. J Basic Microbiol 31:423–428
Coutinho PM, Reilly PJ (1994) Structure-function in the catalytic and starch binding domains of glucoamylase. Protein Eng 7:393–400
Davies E, Larkins BA, Knight RH (1972) Polysomes from peas. An improved method for their isolation in the absence of ribonuclease inhibitors. Plant Physiol 50:581–584
Dohmen RJ, Strasser AWM, Dahlems U, Hollenberg CP (1990) Cloning of the Schwanniomyces occidentalis glucoamylase gene (GAM1) and its expression in Saccharomyces cerevisiae. Gene 95:111–121
Dohmen RJ, Strasser AM, Höner CB, Hollenberg CP (1991) An efficient transformation procedure enabling long term storage of competent cells of various yeast genera. Yeast 7:691–692
Elion EA, Warner JR (1984) The major promoter element of rRNA transcription in yeast lies 1 kb upstream. Cell 39:663–673
Ernst JF (1986) Improved secretion of heterologous proteins by Saccharomyces cerevisiae: effects of promoter substitution in alpha-factor fusions. DNA 5:483–491
Gellissen G, Janowicz ZA, Merckelbach A, Piontek M, Keup P, Weydemann U, Hollenberg CP, Strasser AWM (1991) Heterologous gene expression in Hansenula polymorpha: efficient secretion of glucoamylase. Biotechnology 9:291–295
Gienow U, Kunze G, Schauer F, Bode R, Hofemeister J (1990) The yeast genus Trichosporon spec. Ls3; molecular characterization of genomic complexity. Zentralbl Mikrobiol 145:3–12
Gurr SJ, Unkles SE, Kinghorn JR (1987) The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn JR (ed) Gene structure in eukaryotic microbes. IRL, Oxford, pp 93–139
Hanahan D (1983) Studies on transformation of E. coli with plasmids. J Mol Biol 166:557–580
Heijne G von (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14:4683–4690
Itoh T, Ohtsuki I, Yamashita I, Fukui S (1987) Nucleotide sequence of the glucoamylase gene GLU1 in the yeast Saccharomycopsis fibuligera. J Bacteriol 169:4171–4176
Kunze G., Kunze I (1994) Characterization of Arxula adeninivorans strains from different habitats. Antonie Van Leeuwenhoek 65:29–34
Kunze G, Bode R, Schmidt H, Samsonova IA, Birnbaum D (1987) Identification of a lys2 mutant of Candida maltosa by means of transformation. Curr Genet 11:385–391
Lin WL, Feldberg RS, De Bernardez Clark E (1993) Kinetics of cell growth and heterologous glucoamylase production in recombinant Aspergillus nidulans. Biotechnol Bioeng 41:273–279
Middelhoven WJ, Hoogkamer-Te Niet MC, Kreger-Van Rij NJW (1984) Trichosporon adeninovorans sp. nov., a yeast species utilizing adenine, xanthine, uric acid, putrescine and primary n-alkylamines as the sole source of carbon, nitrogen and energy. Antonie Van Leeuwenhoek 50:369–378
Rose MD, Winston F, Hieter P (1990) Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Sherman F, Fink GR, Hicks JB (1986) Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Smith DE, Fisher PA (1984) Identification, developmental regulation, and response to heat shock of two autogenically related forms of a major nuclear envelope protein in Drosophila. J Cell Biol 99:20–28
Stoltenburg R, Wartmann T, Kunze I, Kunze G (1995) A reliable method to separate free and membrane-bound polysomes from different yeast species. Biotechn 18:564–568
Svensson B, Jespersen H, Sierks MR, MacGregor EA (1989) Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem J 264:309–311
Tubb RS (1986) Amylolytic yeasts for commercial applications. Trends Biotechnol 4:98–104
Van der Walt JP, Smith MT, Yamada Y (1990) Arxula gen. nov. (Candidaceae), a new anamorphic, arthroconidial yeast genus. Antonie Van Leeuwenhoek 57:59–61
Vihinen M, Mäntsälä M (1989) Microbial amylolytic enzymes. Crit Rev Biochem Mol Biol 24:239–418
Yamashita I (1989) The threonine- and serine-rich tract of the secretory glucoamylase can direct β-galactosidase to the cell envelope. Agric Biol Chem 53:483–489
Yamashita I, Suzuki K, Fukui S (1985) Nucleotide sequence of the extracellular glucoamylase gene ST A1 in the yeast Saccharomyces diastaticus. J Bacteriol 161:567–573
Young RA, Davis RW (1983) Yeast RNA polymerase II genes: Isolation with antibody probes. Science 222:778–782
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bui, D.M., Kunze, I., Förster, S. et al. Cloning and expression of an Arxula adeninivorans glucoamylase gene in Saccharomyces cerevisiae . Appl Microbiol Biotechnol 44, 610–619 (1996). https://doi.org/10.1007/BF00172493
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00172493