Summary
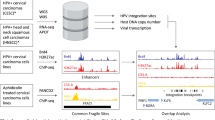
To evaluate the trend of viral integration in the human genome, chromosomal localization of five DNA-containing viruses compiled from literature data was compared to the location of fragile sites and protooncogenes. A total of 35 regionally mapped viral integration sites from tumors and transformed cells were distributed over 19 chromosomes. Of the 35 integration sites 23 (66%) were at the bands of fragile sites, and 7 were one band away (20%). This statistically defines the correlation as highly significant (P = 0.0000183, Fisher's F-test). Five integration sites did not correspond to the location of a fragile site. Thirteen integration sites and proto-oncogenes mapped at the same bands (37%), 6. (17%) were one band apart, and at 16 integration sites (46%) no proto-oncogenes were localized (P = 0.00491). Eighteen viral integration sites, fragile sites, and protooncogenes (51%) were localized at the same bands or one band distant. This clustering of viral integration sites, fragile sites, and proto-oncogenes is statistically highly significant (P = 0.0000118), and indicates nonrandom viral integration in the human genome.
Similar content being viewed by others
References
Alhadeff B, Purrello M, Romani M, Szabo P, Gluzman T, Siniscalco M (1987) The adenovirus 12 modification site 1p(A1ZMZ) is also a site of preferential viral integration in human cells transfected with the hybrid recombinant virus adenovirus 5/ SV40 (abstract). (9th International Workshop on Human Gene Mapping) Cytogenet Cell Genet 46:569
Ambros PF, Karlic HI (1987) Chromosomal insertion of human papillomavirus 18 sequences in HeLa cells detected by non-isotopic in situ hybridization and reflection contrast microscopy. Hum Genet 77:251–254
Bishop JM (1987) The molecular genetics of cancer. Science 235:305–311
Bowcock AM, Pinto MR, Bey E, Kuyl JM, Dusheiko GM, Bernstein R (1985) The PLC/PRF/5 human hepatoma cell line. II. Chromosomal assignment of hepatitis B virus integration sites. Cancer Genet Cytogenet 18:19–26
Cannizzaro LA, Durst M, Mendez MJ, Hecht BK, Hecht F (1988) Regional chromosome localization of human papillomavirus integration sites near fragile sites, oncogenes and cancer chromosome breakpoints. Cancer Genet Cytogenet 33:93–98
Croce C (1981) Integration of oncogenic viruses in mammalian cells. Int Rev Cytol 71:1–17
Dejean AL, Bougueleret L, Grzeschik KM, Tiollais P (1986) Hepatitis B virus DNA integration in a sequence homologous to v-erbA and steroid receptor genes in a hepatocellular carcinoma. Nature 322:70–72
Durnam DM, Menninger JC, Chandler SH, Smith PP, McDougall J (1988) A fragile site in the human U2 small nuclear RNA gene cluster is revealed by adenovirus type 12 infection. Mol Cell Biol 8:1853–1867
Durst M, Croce CM, Gissmann L, Schwarz E, Huebner K (1987) Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc Natl Acad Sci USA 84:1070–1074
Hecht F (1988a) Fragile sites, cancer chromosome breakpoints and oncogenes all cluster in light G-bands. Cancer Genet Cytogenet 31:17–24
Hecht F (1988b) Fragile site update (Appendix A). Cancer Genet Cytogenet 31:125–128
Henderson A (1983) Localization of viral-specific DNA in the mammalian chromosome complement by cytological hybridization. In: Rowley J, Ultman J (eds) Chromosomes and cancer: from molecules to man. Academic Press, New York London, pp 247–272
Henderson A, Ripley S, Heller M, Kieff E (1983) Human chromosome association of Epstein-Barr virus DNA in a Burkitt tumor cell line and in lymphocytes growth-transformed in vitro. Proc Natl Acad Sci USA 80:1987–1991
Henderson A, Ripley S, Hino O, Rogler CE (1988) Identification of a chromosomal aberration associated with a hepatitis B DNA integration site in human cells. Cancer Genet Cytogenet 30:269–275
Hino O, Shows T, Rogler C (1986) Hepatitis B virus integration site in hepatocellular carcinoma at chromosome 17:18 translocation. Proc Natl Acad Sci USA 83:8338–8342
Human Gene Mapping 9 (1987) 9th International Workshop on Human Gene Mapping. Cytogenet Cell Genet 46:1–762
Human Gene Mapping 9.5 (1989) Update to the 9th International Workshop on Human Gene Mapping. Cytogenet Cell Genet 49:1–258
Lazo P, DiPaolo JA, Popescu NC (1989) Amplification of viral transforming genes of human papillomavirus-18 and its 5′ flanking sequences located near myc proto-oncogene in HeLa cells. Cancer Res 49:4305–4310
LeBeau MM (1988) Chromosomal fragile sites and cancer-specific breakpoints — a moderating viewpoint. Cancer Genet Cytogenet 31:55–61
Minceva A, Gissmann L, Hausen H zur (1987) Chromosomal integration sites of human papillomaviruses DNA in three cervical cancer cell lines mapped by in situ hybridization. Med Microbiol Immunol 176:245–256
Mitelman F (1988) Catalog of chromosome aberrations in cancer. Liss, New York
Mitelman F, Heim S (1988) Consistent involvement of only 71 of the 329 chromosomal bands of the human genome in primary neoplasia-associated rearrangements. Cancer Res 48:7115–7119
Nagaya T, Nakamura T, Tokino T, Tsurimoto T, Imai M, Mayumi T, Kamino K, Yamamura K, Matsubara K (1987) The mode of hepatitis B virus DNA integration in chromosomes of human hepatocellular carcinoma. Genes Dev 1:773–782
Pfister H (1987) Human papillomaviruses and genital cancer. Adv Cancer Res 48:113–147
Popescu NC, DiPaolo JA, Amsbaugh SC (1987a) Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet Cell Genet 44:58–62
Popescu NC, Amsbaugh SC, DiPaolo JA (1987b) Human papillomavirus type 18 DNA is integrated at a single chromosome site in cervical carcinoma cell line SW756. J Virol 51:1682–1685
Rabin M, Uhlenbeck OC, Steffensen DM, Mangel WF (1984) Chromosomal sites of integration of Simian virus 40 DNA sequences mapped by in situ hybridization in two transformed hybrid cell lines. J Virol 49:445–451
Robinson HL, Gagnon G (1986) Patterns of proviral insertion and deletion in avian leukosis virus-induced lymphomas. J Virol 57:28–36
Rogler CE, Sherman M, Su CY, Shafritz DA, Summers J, Shows TB, Henderson A, Kew M (1985) Deletion in chromosome 11p associated with a hepatitis B integration site in hepatocellular carcinoma. Science 230:319–322
Rohdewohld H, Weiher H, Reik W, Jaenisch R, Breindl M (1987) Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNaselhypersensitive sites. J Virol 61:336–343
Sandberg AA (1980) Chromosomes in human cancer and leukemia. Elsevier/North-Holland, New York Amsterdam
Shaul Y, Garcia PD, Schonberg S, Rutter WJ (1986) Integration of hepatitis B virus DNA in chromosome-specific satellite sequences. J Virol 59:731–734
Shiraishi Y, Taguchi T, Ohta Y, Hirai K (1985) Chromosomal localization of the Epstein-Barr virus (EBV) genome in Bloom's syndrome B-lymphoblastoid cell lines transformed with EBV. Chromosoma 93:157–164
Sutherland GR, Simmers RN (1988) No statistical association between common fragile sites and nonrandom chromosome breakpoints in cancer cells. Cancer Genet Cytogenet 31:9–15
Tokino T, Kukushige S, Nakamura T, Nagaya T, Murotsu T, Shiga L, Aoki N, Matsubara K (1987) Chromosomal translocation and inverted duplication associated with integrated hepatitis B virus in hepatocellular carcinomas. J Virol 61:3848–3854
Trescol-Biemont MC, Biemont C, Daillie J (1987) Localization polymorphism of EBV DNA genomes in the chromosomes of Burkitt lymphoma cell lines. Chromosoma 95:144–147
Vijaya S, Steffen D, Robinson HL (1986) Acceptor sites for retroviral integrations map near DNaseI-hypersensitive sites in chromatin. J Virol 60:683–692
Wang HP, Rogler CE (1988) Deletions in human chromosome arms 11p and 13q in primary hepatocellular carcinomas. Cytogenet Cell Genet 48:72–78
Weinberg RA (1980) Integrated genomes of animal viruses. Annu Rev Biochem 49:197–226
Yunis JJ, Soreng AL, Bowe AE (1987) Fragile sites are targets of diverse mutagens and carcinogens. Oncogene 1:59–69
Zhou YZ, Slagle BL, Donehower LA, Vantuinen P, Ledbetter DH, Butel JS (1988) Structural analysis of a hepatitis B virus genome integrated into chromosome 17p of a human hepatocellular carcinoma. J Virol 62:4224–4231
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Popescu, N.C., Zimonjic, D. & DiPaolo, J.A. Viral integration, fragile sites, and proto-oncogenes in human neoplasia. Hum Genet 84, 383–386 (1990). https://doi.org/10.1007/BF00195804
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00195804