Summary
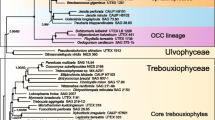
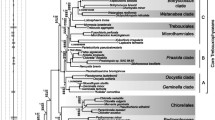
Complete small-subunit rRNA (16S-like rRNA) coding region sequences were determined for eight species of the Chlorococcales (Chlorophyceae). The genera investigated includePrototheca, Ankistrodesmus, Scenedesmus, and fiveChlorella species. Distance matrix methods were used to infer a phylogenetic tree that describes evolutionary relationships between several plant and green algal groups. The tree exhibits a bifurcation within the Chlorococcales consistent with the division into Oocystaceae and Scenedesmaceae, but three of the fiveChlorella species are more similar to other algae than toChlorella vulgaris. All of the sequences contain primary and secondary structural features that are characteristic of 16S-like rRNAs of chlorophytes and higher plants.Anikstrodesmus stipitatus, however, contains a 394-bp group I intervening sequence in its 16S-like rRNA coding region.
Similar content being viewed by others
References
Atmadja J, Brimacombe R, Maden BEH (1984)Xenopus laevis 18S ribosomal RNA: experimental determination of secondary structural elements, and locations of methyl groups in the secondary structure model. Nucleic Acids Res 12:2649–2667
Bold HC, Wynne MJ (1985) Introduction to the algae. Prentice-Hall, Englewood Cliffs NJ
Cech TR (1988) Conserved sequences and structures of group I introns: building an active site for RNA catalysis—a review. Gene 73:259–271
Choi YC (1985) Structural organization of ribosomal RNAs from Novikoff hepatoma. I. Characterization of fragmentation products from 40S subunit. J Biol Chem 260:12769–12772
Dams E, Hendriks L, Van de Peer Y, Neefs JM, Smits G, Vandenbempt I, De Wachter R (1988) Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res 16(suppl):r87-r173
Eckenrode VK, Arnold J, Meagher RB (1985) Comparison of the nucleotide sequence of soybean 18S rRNA with the sequences of other small-subunit rRNAs. J Mol Evol 21:259–269
Elwood HJ, Olsen GJ, Sogin ML (1985) The small-subunit ribosomal RNA gene sequences from the hypotrichous ciliatesOxytricha nova andStylonychia pustulata. Mol Biol Evol 2: 399–410
Felsenstein J (1988) Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet 22:521–565
Fitch WM, Margoliash E (1967) Construction of phylogenetic trees. Science 155:279–284
Fott B, Lochhead R, Clémençon H (1975) Taxonomie der ArtenChlorella ultrasquamata Clém. et Fott undChlorella fusca Shih. et Krauss. Arch Protistenkd 117:288–296
Fott B, Nováková M (1969) A monograph of the genusChlorella. The fresh water species. In: Fott B (ed) Studies in phycology. Academia, Prague, pp 10–74
Gonzalez IL, Schmickel RD (1986) The human 18S ribosomal RNA gene: evolution and stability. Am J Hum Genet 38: 419–427
Götz H, Arnold CG (1980a) Comparative electrophoretic study on ribosomal proteins from algae. Planta 149:19–26
Götz H, Arnold CG (1980b) Analysis of ribosomal proteins from various species of algae. Comparative electrophoretic study on proteins from chloroplast ribosomes. Biochem Physiol Pflanz 175:1–8
Gunderson JH, Sogin ML (1986) Length variation in eukaryotic rRNAs: small-subunit rRNAs from the protistsAcanthamoeba castellanii andEuglena gracilis. Gene 44:63–70
Gunderson JH, Elwood HJ, Ingold A, Kindle K, Sogin ML (1987) Phylogenetic relationships between chlorophytes, chrysophytes and oomycetes. Proc Natl Acad Sci USA 84:5823–5827
Gutell RR, Weiser B, Woese CR, Noller HF (1985) Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol 32:155–216
Hegewald E (1982) Taxonomisch-morphologische Untersuchung vonScenedesmus-Isolaten aus Stammsarmmlungen. Algol Stud 29:375–406
Hellmann V, Kessler E (1974) Physiologische und biochemische Beiträge zur Taxonomie der GattungChlorella. VIII. Die Basenzusammensetzung der DNS. Arch Microbiol 95: 311–318
Hendriks L, De Baere R, Van Broeckhoven C, De Wachter R (1988) Primary and secondary structure of the 18S ribosomal RNA of the insect speciesTenebrio molitor. FEBS Lett 232: 115–120
Herzog M, Maroteaux L (1986) Dinoflagellate 17S rRNA sequence inferred from the gene sequence: evolutionary implications. Proc Natl Acad Sci USA 83:8644–8648
Hindák F (1982) Taxonomic position of the chlorococcal algaChlorella zofingiensis Dönz 1934 (Chlorophyceae). Algol Stud 30:13–23
Huss VAR, Sogin ML (1989) Primary structure of theChlorella vulgaris small subunit RNA coding region. Nucleic Acids Res 17:1255
Huss VAR, Dörr R, Grossmann U, Kessler E (1986) Deoxyribonucleic acid reassociation in the taxonomy of the genusChlorella. I.Chlorella sorokiniana. Arch Microbiol 145:329–333
Huss VAR, Schwarzwälder E, Kessler E (1987a) Deoxyribonucleic acid reassociation in the taxonomy of the genusChlorella. II.Chlorella saccharophila. Arch Microbiol 147:221–224
Huss VAR, Hehenberger A, Kessler E (1987b) Deoxyribonucleic acid reassociation in the taxonomy of the genusChlorella. III.Chlorella fusca andChlorella kessleri. Arch Microbiol 149:1–3
Huss VAR, Wein KH, Kessler E (1988) Deoxyribonucleic acid reassociation in the taxonomy of the genusChlorella. IV.Chlorella protothecoides and its relationship to the genusPrototheca. Arch Microbiol 150:509–511
Huss VAR, Huss G, Kessler E (1989a) Deoxyribonucleic acid reassociation and interspecies relationships of the genusChlorella (Chlorophyceae). Plant Syst Evol 168:71–82
Huss VAR, Scharpf TK, Kessler E (1989b) Deoxyribonucleic acid reassociation in the taxonomy of the genusChlorella. V.Chlorella vulgaris, C. luteoviridis, C. minutissima, andC. zofingiensis. Arch Microbiol 152:512–514
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kalina T, Punčochářová M (1987) Taxonomy of the subfamily Scotiellocystoideae Fott 1976 (Chlorellaceae, Chlorophyceae). Algol Stud 45:473–521
Kerfin W, Kessler E (1978) Physiological and biochemical contributions to the taxonomy of the genusPrototheca. II. Starch hydrolysis and base composition of DNA. Arch Microbiol 116:105–107
Kessler E (1976) Comparative physiology, biochemistry, and the taxonomy ofChlorella (Chlorophyceae). Plant Syst Evol 125:129–138
Kessler E (1980) Physiological and biochemical contributions to the taxonomy of the generaAnkistrodesmus andScenedesmus. V. Starch hydrolysis and new assignment of strains. Arch Microbiol 126:11–14
Kessler E (1982a) Chemotaxonomy in the Chlorococcales. In: Round FE, Chapman DJ (eds) Progress in phycological research, vol. 1. Elsevier Biomedical Press BV, Amsterdam, pp 111–135
Kessler E (1982b) Physiological and biochemical contributions to the taxonomy of the genusPrototheca. III. Utilization of organic carbon and nitrogen compounds. Arch Microbiol 132: 103–106
Kessler E (1984) A general review on the contribution of chemotaxonomy to the systematics of green algae. In: Irvine DEG, John DM (eds) Systematics of the green algae. Academic Press, London, pp 391–407
Kessler E (1987) Separation ofChlorella ellipsoidea fromC. saccharophila (Chlorophyceae): no growth on mannitol and cadmium sensitivity. Plant Syst Evol 157:247–251
Kiss T, Szkukálek A, Solymosy F (1989) Nucleotide sequence of a 17S (18S) rRNA gene from tomato. Nucleic Acids Res 17:2127
Komárek J (1987) Species concept in coccal green algae. Algol Stud 45:437–471
Kümmel H, Kessler E (1980) Physiological and biochemical contributions to the taxonomy of the genusChlorella. XIII. Serological studies. Arch Mirobiol 126:15–19
Medlin L, Elwood HJ, Stickel S, Sogin ML (1988) The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499
Menzel K, Wild A (1989) A comparative ultrastructural investigation of someNannochloris species (Chlorococcales) with particular reference to the systematic position ofNanochlorum eucaryotum. Botanica Acta 102:152–158
Messing J (1983) New M13 vectors for cloning. Methods Enzymol 101:20–78
Messing J, Carlson J, Hagen G, Rubenstein I, Oleson A (1984) Cloning and sequencing of the ribosomal RNA genes in maize: the 17S region. DNA 3:31–40
Nadakuvakaren MJ, McCracken DA (1973)Prototheca: an alga or a fungus? J Phycol 9:113–116
Nairn CJ, Ferl RJ (1988) The complete nucleotide sequence of the small-subunit ribosomal RNA coding region for the cycadZamia pumila: phylogenetic implications. J Mol Evol 27: 133–141
Patterson GW (1974) Sterols of some green algae. Comp Biochem Physiol 47B:453–457
Pore RS (1972) Nutritional basis for relatingPrototheca andChlorella. Can J Microbiol 18:1175–1177
Pore RS (1985)Prototheca taxonomy. Mycopathologia 90:129–139
Pore RS, Barnett EA, Barnes WC Jr, Walker JD (1983)Prototheca ecology. Mycopathologia 81:49–62
Raué HA, Klootwijk J, Musters W (1988) Evolutionary conservation of structure and function of high molecular weight ribosomal RNA. Prog Biophys Mol Biol 51:77–129
Rausch H, Larsen N, Schmitt R (1989) Phylogenetic relationships of the green algaVolvox carteri deduced from smallsubunit ribosomal RNA comparisons. J Mol Evol 29:255–265
Rubtsov PM, Musakhanov MM, Zakharyev VM, Krayev AS, Skryabin KG, Bayev AA (1980) The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene fromSaccharomyces cerevisiae. Nucleic Acids Res 8:5779–5794
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491
Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94:441–448
Sargent M, Zahn R, Walters B, Gupta R, Kaine B (1988) Nucleotide sequence of the 18S rDNA from the microalgaNanochlorum eucaryotum. Nucleic Acids Res 16:4156
Shihira I, Krauss RW (1965)Chlorella. Physiology and taxonomy of forty-one isolates. University of Maryland, College Park
Soeder CJ (1980) Massive cultivation of microalgae: results and prospects. Hydrobiologia 72:197–209
Sogin ML, Ingold A, Karlok M, Nielsen H, Engberg J (1986) Phylogenetic evidence for the acquisition of ribosomal RNA introns subsequent to the divergence of some of the majorTetrahymena groups. EMBO J 5:3625–3630
Sogin ML, Gunderson JH, Elwood HJ, Rogelio AA, Peattie DA (1989) Phylogenetic meaning of the kingdom, concept: an unusual ribosomal RNA fromGiardia lamblia. Science 243: 75–77
Sudman MS (1974) Protothecosis. Am J Clin Pathol 61:10–19
Takaiwa F, Oono K, Sugiura M (1984) The complete nucleotide sequence of a rice 17S rRNA gene. Nucleic Acids Res 12: 5441–5448
Walker JD, Colwell RR, Vaituzis Z, Meyer SA (1975) Petroleum-degrading achlorophyllous algaPrototheca zopfii. Nature 254:423–424
Walker JD, Pore RS (1978) Growth ofPrototheca isolated on n-hexadecane and mixed hydrocarbon substrate. Appl Environ Microbiol 35:694–697
Wilhelm C, Eisenbeis G, Wild A, Zahn R (1982)Nanochlorum eucaryotum: a very reduced coccoid species of marine Chlorophyceae. Z Naturforsch 37c:107–114
Woese CR, Gutell R, Gupta R, Noller HF (1983) Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev 47:621–669
Woese CR, Fox E (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA 74:5088–5090
Zahn RK (1984) A green alga with minimal eukaryotic features:Nanochlorum eucaryotum. Origins Life 13:289–303
Zuckerkandl E, Pauling L (1965) Molecules as documents of evolutionary history. J Theor Biol 8:357–366
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Huss, V.A.R., Sogin, M.L. Phylogenetic position of someChlorella species within the chlorococcales based upon complete small-subunit ribosomal RNA sequences. J Mol Evol 31, 432–442 (1990). https://doi.org/10.1007/BF02106057
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02106057