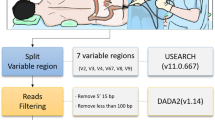
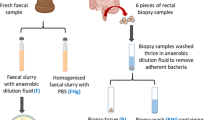
Abstract
Limited data exist on the spatial distribution of the colonic bacteria in humans. We collected the colonic biopsies from five segments of 27 polyp-free adults and collected feces from 13 of them. We sequenced the V4 region of the bacterial 16S rRNA gene using the MiSeq platform. The sequencing data were assigned to the amplicon sequence variant (ASV) using SILVA. Biodiversity and the relative abundance of the ASV were compared across the colonic segments and between the rectal and fecal samples. Bacterial functional capacity was assessed using Tax4fun. Each individual had a unique bacterial community composition (Weighted Bray–Curtis P value = 0.001). There were no significant differences in richness, evenness, community composition, and the taxonomic structure across the colon segments in all the samples. Firmicutes (47%), Bacteroidetes (39%), and Proteobacteria (6%) were the major phyla in all segments, followed by Verrucomicrobia, Fusobacteria, Desulfobacterota, and Actinobacteria. There were 15 genera with relative abundance > 1%, including Bacteroides, Faecalibacterium, Escherichia/Shigella, Sutterella, Akkermansia, Parabacteroides, Prevotella, Lachnoclostridium, Alistipes, Fusobacterium, Erysipelatoclostridium, and four Lachnospiraceae family members. Intra-individually, the community compositional dissimilarity was the greatest between the cecum and the rectum. There were significant differences in biodiversity and the taxonomic structure between the rectal and fecal bacteria. The bacterial community composition and structure were homogeneous across the large intestine in adults. The inter-individual variability of the bacteria was greater than inter-segment variability. The rectal and fecal bacteria differed in the community composition and structure.






Similar content being viewed by others
Data Availability
Data will be available at NCBI.
References
Lozupone CA (2016) Getting to know the microbiome. Nat Microbiol 1:16030. https://doi.org/10.1038/nmicrobiol.2016.30
Liu S, Zhao W, Lan P, Mou X (2020) The microbiome in inflammatory bowel diseases: from pathogenesis to therapy. Protein Cell. https://doi.org/10.1007/s13238-020-00745-3
Meng X, Zhang G, Cao H, Yu D, Fang X, de Vos WM, Wu H (2020) Gut dysbacteriosis and intestinal disease: mechanism and treatment. J Appl Microbiol 129(4):787–805. https://doi.org/10.1111/jam.14661
Feldman M, Friedman LS, Brandt LJ (2010) Sleisenger and fordtran's gastrointestinal and liver disease E-Book: pathophysiology, diagnosis, management, expert consult premium edition-enhanced online features, vol 1. Elsevier Health Sciences
Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, Wu GD (2014) Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147(5):1055-1063 e1058
O’May GA, Reynolds N, Smith AR, Kennedy A, Macfarlane GT (2005) Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. J Clin Microbiol 43(7):3059–3065. https://doi.org/10.1128/JCM.43.7.3059-3065.2005
Tropini C, Earle KA, Huang KC, Sonnenburg JL (2017) The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21(4):433–442. https://doi.org/10.1016/j.chom.2017.03.010
Li D, Chen H, Mao B, Yang Q, Zhao J, Gu Z, Zhang H, Chen YQ, Chen W (2017) Microbial biogeography and core microbiota of the rat digestive tract. Sci Rep 8:45840. https://doi.org/10.1038/srep45840
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308(5728):1635–1638. https://doi.org/10.1126/science.1110591
Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD (2011) Bacterial biogeography of the human digestive tract. Sci Rep 1:170. https://doi.org/10.1038/srep00170
Green GL, Brostoff J, Hudspith B, Michael M, Mylonaki M, Rayment N, Staines N, Sanderson J, Rampton DS, Bruce KD (2006) Molecular characterization of the bacteria adherent to human colorectal mucosa. J Appl Microbiol 100(3):460–469. https://doi.org/10.1111/j.1365-2672.2005.02783.x
Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI (2011) Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS ONE 6(9):e25042. https://doi.org/10.1371/journal.pone.0025042
Lepage P, Seksik P, Sutren M, de la Cochetiere MF, Jian R, Marteau P, Dore J (2005) Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis 11(5):473–480
Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM (2002) Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 68(7):3401–3407
Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X, Ma ZS, Shi P (2014) Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J 8(4):881–893. https://doi.org/10.1038/ismej.2013.185
Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR (2012) Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J 6(1):57–70. https://doi.org/10.1038/ismej.2011.90
Liu Y, Ajami NJ, El-Serag HB, Hair C, Graham DY, White DL, Chen L, Wang Z, Plew S, Kramer J, Cole R, Hernaez R, Hou J, Husain N, Jarbrink-Sehgal ME, Kanwal F, Ketwaroo G, Natarajan Y, Shah R, Velez M, Mallepally N, Petrosino JF, Jiao L (2019) Dietary quality and the colonic mucosa-associated gut microbiome in humans. Am J Clin Nutr 110(3):701–712. https://doi.org/10.1093/ajcn/nqz139
American Society of C, Rectal S, American Society for Gastrointestinal E, Society of American G, Endoscopic S, Wexner SD, Beck DE, Baron TH, Fanelli RD, Hyman N, Shen B, Wasco KE (2006) A consensus document on bowel preparation before colonoscopy: prepared by a Task Force from the American Society of Colon and Rectal Surgeons (ASCRS), the American Society for Gastrointestinal Endoscopy (ASGE), and the Society of American Gastrointestinal and Endoscopic Surgeons (SAGES). Surg Endosc 20(7):1161. https://doi.org/10.1007/s00464-006-3037-1
Human Microbiome Project C (2012) A framework for human microbiome research. Nature 486(7402):215–221
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6(8):1621–1624. https://doi.org/10.1038/ismej.2012.8
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998. https://doi.org/10.1038/nmeth.2604
Edgar RC (2018) Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ 6:e4652. https://doi.org/10.7717/peerj.4652
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41((Database issue)):D590-596. https://doi.org/10.1093/nar/gks1219
Caruso V, Song X, Asquith M, Karstens L (2019) Performance of microbiome sequence inference methods in environments with varying biomass. mSystems 4(1). https://doi.org/10.1128/mSystems.00163-18
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13(7):581–583. https://doi.org/10.1038/nmeth.3869
Asshauer KP, Wemheuer B, Daniel R, Meinicke P (2015) Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31(17):2882–2884. https://doi.org/10.1093/bioinformatics/btv287
Dethlefsen L, Eckburg PB, Bik EM, Relman DA (2006) Assembly of the human intestinal microbiota. Trends Ecol Evol 21(9):517–523. https://doi.org/10.1016/j.tree.2006.06.013
Zhu A, Sunagawa S, Mende DR, Bork P (2015) Inter-individual differences in the gene content of human gut bacterial species. Genome Biol 16:82. https://doi.org/10.1186/s13059-015-0646-9
Lampe JW, Navarro SL, Hullar MA, Shojaie A (2013) Inter-individual differences in response to dietary intervention: integrating omics platforms towards personalised dietary recommendations. Proc Nutr Soc 72(2):207–218. https://doi.org/10.1017/S0029665113000025
Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488(7410):178–184. https://doi.org/10.1038/nature11319
Lyra A, Forssten S, Rolny P, Wettergren Y, Lahtinen SJ, Salli K, Cedgard L, Odin E, Gustavsson B, Ouwehand AC (2012) Comparison of bacterial quantities in left and right colon biopsies and faeces. World J Gastroenterol 18(32):4404–4411. https://doi.org/10.3748/wjg.v18.i32.4404
Neish AS (2014) Mucosal immunity and the microbiome. Ann Am Thorac Soc 11(Suppl 1):S28-32. https://doi.org/10.1513/AnnalsATS.201306-161MG
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105(43):16731–16736. https://doi.org/10.1073/pnas.0804812105
Chen W, Liu F, Ling Z, Tong X, Xiang C (2012) Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 7(6):e39743. https://doi.org/10.1371/journal.pone.0039743
Peng C, Ouyang Y, Lu N, Li N (2020) The NF-kappaB signaling pathway, the microbiota, and gastrointestinal tumorigenesis: recent advances. Front Immunol 11:1387. https://doi.org/10.3389/fimmu.2020.01387
Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A (2020) The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol 11:906. https://doi.org/10.3389/fimmu.2020.00906
Yamauchi M, Lochhead P, Morikawa T, Huttenhower C, Chan AT, Giovannucci E, Fuchs C, Ogino S (2012) Colorectal cancer: a tale of two sides or a continuum? Gut 61(6):794–797. https://doi.org/10.1136/gutjnl-2012-302014
Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC (2013) Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointestinal Liver Physiol 305(5):G341-347. https://doi.org/10.1152/ajpgi.00046.2013
Durban A, Abellan JJ, Jimenez-Hernandez N, Ponce M, Ponce J, Sala T, D’Auria G, Latorre A, Moya A (2011) Assessing gut microbial diversity from feces and rectal mucosa. Microb Ecol 61(1):123–133. https://doi.org/10.1007/s00248-010-9738-y
Acknowledgements
We thank Jocelyn Uriostegui, Sarah Plew, Ashley Johnson, Preksha Shah, Kathryn Royse, Ava Smith, and Mahmoud Al-Saadi for patient recruitment. We are grateful for Dr. David Ramsey’s help with data management and Professor Antone Opekun’s assistance with the study.
Funding
This research is supported in part by the Gillson Longenbaugh Foundation and Golfers Against Cancer organization (to LJ), the Cancer Prevention Research Institute of Texas (CPRIT) (RP#140767, to LJ and JFP), the National Institute of Diabetes and Digestive and Kidney Disease P30DK56338 (to HBE), the Cancer Center Support Grant NIH:NCI P30CA022453, and the Houston VA HSR&D Center for Innovations in Quality, Effectiveness and Safety (CIN13-413), and the Alkek research fund (to JFP). Dr. Jiao received partial salary support from National Cancer Institute R01CA172880 (to LJ). Dr. White receives partial salary support from the Department of Veterans Affairs (I01CX001430). The opinions expressed reflect those of the authors and not necessarily those of the Department of Veterans Affairs, the US government, or Baylor College of Medicine. The funding sources had no role in the study design, data collection, analysis, interpretation, and writing.
Author information
Authors and Affiliations
Contributions
Li Jiao was responsible for the design, data analysis, and writing. Joseph F. Petrosino formed the study concept and was responsible for microbiota profiling and bioinformatics. Themistoklis Kourkoumpetis, and Diane Hutchinson participated in writing the manuscript. Diane Hutchinson, Nadim J. Ajami, and Kristi Hoffman managed the project and performed the bioinformatics analysis. Donna L. White and Jennifer Kramer participated in patient enrollment and edited the manuscript; David Y. Graham and Hashem B. El-Serag managed the clinic and edited the manuscript. Clark Hair, Rajesh Shah, Fasiha Kanwal, Maria Jarbrink-Sehgal, Nisreen Husain, Ruben Hernaez, Jason Hou, Rhonda Cole, Maria Velez, and Gyanprakash Ketwaroo collected specimens and edited the manuscript. All authors approved the content of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The study protocol (#H30941) was approved by the Institutional Review Board of Baylor College of Medicine and Michael E. DeBakey VA Medical Center.
Consent to Participate
A trained research coordinator obtained informed consent from each participant.
Consent for Publication
Participants provided informed consent to publish the research data in aggregate.
Conflict of Interest
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiao, L., Kourkoumpetis, T., Hutchinson, D. et al. Spatial Characteristics of Colonic Mucosa-Associated Gut Microbiota in Humans. Microb Ecol 83, 811–821 (2022). https://doi.org/10.1007/s00248-021-01789-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01789-6