Abstract
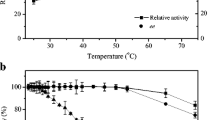
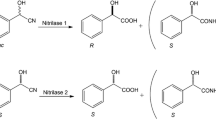
(R)-mandelic acid was produced from racemic mandelonitrile using free and immobilized cells of Pseudomonas putida MTCC 5110 harbouring a stereoselective nitrilase. In addition to the optimization of culture conditions and medium components, an inducer feeding approach is suggested to achieve enhanced enzyme production and therefore higher degree of conversion of mandelonitrile. The relationship between cell growth periodicity and enzyme accumulation was also studied, and the addition of the inducer was delayed by 6 h to achieve maximum nitrilase activity. The nitrilase expression was also authenticated by the sodium dodecyl phosphate-polyacrylamide gel electrophoresis analysis. P. putida MTCC 5110 cells were further immobilized in calcium alginate, and the immobilized biocatalyst preparation was used for the enantioselective hydrolysis of mandelonitrile. The immobilized system was characterized based on the Thiele modulus (ϕ). Efficient biocatalyst recycling was achieved as a result of immobilization with immobilized cells exhibiting 88% conversion even after 20 batch recycles. Finally, a fed batch reaction was set up on a preparative scale to produce 1.95 g of (R)-(-)-mandelic acid with an enantiomeric excess of 98.8%.







Similar content being viewed by others
References
Aksu Z, Bulbul G (1999) Determination of effective diffusion co-efficient of phenol in Ca-alginate immobilized P. putida beads. Enzyme Microb Technol 25:344–348
Banerjee A, Sharma R, Banerjee UC (2002) Nitrile degrading enzymes: current status and future prospects. Appl Microbiol Biotechnol 60:33–44
Banerjee A, Kaul P, Sharma R, Banerjee UC (2003) A high-throughput amenable colorimetric assay for enantioselective screening of nitrilase producing microorganisms. J Biomol Screen 8:559–565
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein, utilizing the principle of protein dye binding. Anal Biochem 72:248–254
Brenner C (2002) Catalysis in the nitrilase superfamily. Curr Opin Chem Biol 12:775–782
Chen D, Lewandowski Z, Roe F, Surapaneni P (1993) Diffusivity of Cu2+ in Ca-alginate gel beads. Biotechnol Bioeng 41:755–760
Goldhust A, Bohak Z (1989) Induction, purification and characterization of the nitrilase of Fusarium oxysporum. Biotechnol Appl Biochem 11:581–601
Inagaki M, Hiratake J, Nishioka T, Oda J (1992) One pot synthesis of optically active cyanohydrin acetates from aldehydes via lipase catalyzed kinetic resolution coupled with in situ formation and racemization of cyanohydrins. J Org Chem 57:5643–5649
Jamuna R, Ramakrishna SV (1992) Continuous synthesis of thermostable α-amylase by Bacillus cells immobilized in Ca-alginate. Enzyme Microb Technol 14:36–41
Kakeya H, Sakai N, Sugai T, Ohta H (1991) Preparation of optically active α-hydroxy acid derivatives by microbial hydrolysis of cyanohydrins and its application to the synthesis of (R)-4-dodecanolide. Agric Biol Chem 55:1877–1881
Kaul P, Banerjee A, Mayilraj S, Banerjee UC (2004) Screening for enantioselective nitrilases: kinetic resolution of racemic mandelonitrile to (R)-(-)-mandelic acid by new bacterial isolates. Tetrahedron Asymmetry 15:207–211
Kobayashi M, Yanaka N, Nagasawa T, Yamada H (1991) Hyperinduction of an aliphatic nitrilase by Rhodococcus rhodochrous K22. FEMS Microbiol Lett 77:121–124
Lammeli UK (1970) Cleavage of structural proteins during the assembly of the head the bacteriophage T4. Nature 227:680–685
Layh N, Stolz A, Forster S, Effenberger F, Knackmuss H-J (1992) Enantioselective hydrolysis of O-acetylmandelonitrile to O-acetylmandelic acid by bacterial nitrilases. Arch Microbiol 158:405–411
Mauger J, Nagasawa T, Yamada H (1990) Occurrence of a novel nitrilase, arylacetonitrilase in Alcaligenes faecalis JM3. Arch Microbiol 155:1–6
Monsan P, Durand G, Navarro JM (1987) Immobilization of microbial cells by adsorption to solid supports. Methods Enzymol 135:307–318
Mylerova V, Martinkova L (2003) Synthetic applications of nitrile converting enzymes. Curr Org Chem 7:1–17
Nagasawa T, Nanba H, Ryuno K, Takeuchi K, Yamada H (1987) Nitrile hydratase of Pseudomonas chlororaphis B23: purification and characterization. Eur J Biochem 162:691–698
Nagasawa T, Kobayashi M, Yamada H (1988) Optimum culture conditions for the production of benzonitrilase by Rhodococcus rhodochrous J1. Arch Microbiol 150:89–94
Nagasawa T, Nakamura T, Yamada H (1990) ɛ-Caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch Microbiol 155:13–17
O’Reilly C, Turner PD (2003) The nitrile family of CN hydrolysing enzymes—a comparative study. J Appl Microbiol 95:1161–1174
Sugai T, Yamazaki T, Yokoyama M, Ohta H (1997) Biocatalysis in organic synthesis: the use of nitrile and amide hydrolysing microorganisms. Biosci Biotechnol Biochem 61:1419–1427
Tanaka H, Matsumura M, Veliky IA (1984) Diffusion characteristics of substrates in Ca-alginate gel beads. Biotechnol Bioeng 26:53–58
Watanabe I, Satoh Y, Enomoto K, Seki S, Sakshita K (1987) Optimal conditions for cultivation of Rhodococcus sp. N774 and conversion of acrylamide by resting cells. Agric Biol Chem 51:3201–3206
Wu ZL, Li ZY (2002) Enhancement of enzyme activity and enantioselectivity via cultivation in nitrile metabolism by Rhodococcus sp. CGMCC 0497. Biotechnol Appl Biochem 35:61–67
Yamada H, Ryuno K, Nagasawa T, Enomoto K, Watanabe I (1986) Optimum culture conditions for production of nitrile hydratase by Pseudomonas chlororaphis B23. Agric Biol Chem 50:2859–2865
Yamamoto K, Komatsu K (1991) Purification and characterization of nitrilase responsible for the hydrolysis from Acinetobacter sp. AK 226. Agric Biol Chem 55:1459–1466
Yamamoto K, Oishi K, Fujimatsu I, Komatsu K (1991) Production of R-(-)- mandelic acid from mandelonitrile by Alcaligenes faecalis ATCC 8750. Appl Environ Microbiol 57:3028–3032
Acknowledgement
A. Banerjee and P. Kaul gratefully acknowledge the fellowship provided by CSIR, Government of India. This is NIFER communication number 346.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at https://doi.org/10.1007/s00253-017-8413-3.
Rights and permissions
About this article
Cite this article
Banerjee, A., Kaul, P. & Banerjee, U.C. Enhancing the catalytic potential of nitrilase from Pseudomonas putida for stereoselective nitrile hydrolysis. Appl Microbiol Biotechnol 72, 77–87 (2006). https://doi.org/10.1007/s00253-005-0255-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0255-8