Abstract
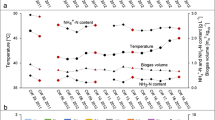
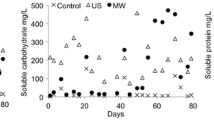
Qualitative and quantitative shifts in methanogen community structure, associated with process performance data, were investigated during the batch anaerobic digestion of a cheese-processing wastewater, whey permeate. Denaturing gradient gel electrophoresis (DGGE) and real-time PCR techniques were applied to obtain qualitative and quantitative microbial data sets, respectively, based on methanogen 16S rRNA genes. Throughout the operation, dynamic variations in both qualitative and quantitative community structures were observed, with repeated shifts in dominance between the aceticlastic Methanosarcinaceae (suggested mainly by the detection of a Methanosarcina-like population) and the hydrogenotrophic Methanomicrobiales (suggested mainly by the detection of a Methanofollis-like population). This trend corresponded well to the diauxic utilization of acetate and longer-chain fatty acids (C3–C6), mainly propionate. Joint-plot non-metric multidimensional scaling (NMS) analysis demonstrated that the qualitative and quantitative community shifts had significant correlations with the composition of residual organic acids and the methane production rate, respectively. This suggests the potential use of microbial community shift analysis as an indicative tool for diagnosing anaerobic digestion processes. The results suggest that more attention should be directed to quantitative, as well as qualitative, approaches for a better understanding of anaerobic digestion, particularly in terms of biogas production efficiency, under dynamic and transitional conditions.





Similar content being viewed by others
References
Akarsubasi AT, Ince O, Kirdar B, Oz NA, Orhon D, Curtis TP, Head IM, Ince BK (2005) Effect of wastewater composition on archaeal population diversity. Water Res 39:1576–1584
Amann RI, Ludwig W, Schleifer K (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Angenent LT, Sung S, Raskin L (2002) Methanogenic population dynamics during startup of a full scale anaerobic sequencing batch reactor treating swine waste. Water Res 36:4648–4654
APHA-AWWA-WEF (2005) Standard methods for the examination of water and wastewater, 21st edn. American Public Health Association, Washington, DC
Boone DR, Castenholz RW (2001) Bergey's manual of systematic bacteriology: the archaea and the deeply branching and phototrophic bacteria, 2nd edn. Springer, New York
Calli B, Mertoglu B, Inanc B, Yenigun O (2005) Methanogenic diversity in anaerobic bioreactors under extremely high ammonia levels. Enzyme Microb Technol 37:448–455
Connaughton S, Collins G, O'Flaherty V (2006) Development of microbial community structure and activity in high-rate anaerobic bioreactor at 18°C. Water Res 40:1009–1017
Fernandez AS, Hashsham SA, Dollhopf SL, Raskin L, Glagoleva O, Dazzo FB, Hickey RF, Criddle CS, Tiedje JM (2000) Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol 66:4058–4067
Forney LJ, Zhou X, Brown CJ (2004) Molecular microbial ecology: land of the one-eyed king. Curr Opin Microbiol 7:210–220
Fry J (2000) Bacterial diversity and 'unculturables'. Microbiol Today 27:186–188
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60–63
Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y (2006) Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl Environ Microbiol 72:1623–1630
Hwang K, Shin SG, Kim J, Hwang S (2008) Methanogenic profiles by denaturing gradient gel electrophoresis using order-specific primers in anaerobic sludge digestion. Appl Microbiol Biotechnol 80:269–276
Hwang SH, Hansen CL (1992) Performance of upflow anaerobic sludge blanket (UASB) reactor treating whey permeate. Trans ASAE 35:1665–1671
Kim J, Lee C, Shin SG, Hwang S (2008) Correlation of microbial mass with ATP and DNA concentrations in acidogenesis of whey permeate. Biodegradation 19:187–195
Klein D (2002) Quantification using real-time PCR technology: applications and limitations. Trend Mol Med 8:257–260
Lee C, Kim J, Shin SG, Hwang S (2008) Monitoring bacterial and archaeal community shifts in a mesophilic anaerobic batch reactor treating a high-strength organic wastewater. FEMS Microbiol Ecol 65:544–554
Lee C, Kim J, Hwang K, O'Flaherty V, Hwang S (2009) Quantitative analysis of methanogenic community dynamics in three anaerobic batch digesters treating different wastewaters. Water Res 43:157–165
McCune B, Grace JB (2002) Analysis of ecological communities. ed. MjM Software Design, Glenden Beach, OR, USA
McMahon KD, Zheng D, Stams AJM, Mackie RI, Raskin L (2004) Microbial population dynamics during start-up and overload conditions of anaerobic digesters treating municipal solid waste and sewage sludge. Biotechnol Bioeng 87:823–834
Muyzer G, Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L (2005) Bacterial population active in metabolism of C1 compounds in the sediment of Lake Washington, a fresh water lake. Appl Environ Microbiol 71:6885–6899
O'Flaherty V, Colohan S, Mulkerrins D, Colleran E (1999) Effect of sulphate addition on volatile fatty acid and ethanol degradation in an anaerobic hybrid reactor. II: microbial interactions and toxic effects. Bioresour Technol 68:109–120
Obata K, Segawa O, Yakabe M, Ishida Y, Kuroita T, Ikeda K, Kawakami B, Kawamura Y, Yohda M, Matsunaga T, Tajima H (2001) Development of a novel method for operating magnetic particles, magtration technology, and its use for automating nucleic acid purification. J Biosci Bioeng 91:500–503
Raskin L, Zheng D, Griffin ME, Stroot PG, Misra P (1995) Characterization of microbial communities in anaerobic bioreactors using molecular probes. Antonie Leeuwenhoek 68:297–308
Roest K, Heilig HGHJ, Smidt H, de Vos WM, Stams AJM, Akkermans ADL (2005) Community analysis of a full-scale anaerobic bioreactor treating paper mill wastewater. Syst Appl Microbiol 28:175–185
Sawayama S, Tsukahara K, Yagishita T (2006) Phylogenetic description of immobilized methanogenic community using real-time PCR in a fixed-bed anaerobic digester. Bioresour Technol 97:69–76
Shin SG, Lee C, Hwang K, Ahn J-H, Hwang S (2008) Use of order-specific primers to investigate the methanogenic diversity in acetate enrichment system. J Ind Microbiol Biotechnol 35:1345–1352
Show K-Y, Wang Y, Foong S-F, Tay JHJ-H (2004) Accelerated start-up and enhanced granulation in upflow anaerobic sludge blanket reactors. Water Res 38:2293–2304
Stams AJM, Dijkema C, Plugge CM, Lens P (1998) Contribution of 13C-NMR spectroscopy to the elucidation of pathways of propionate formation and degradation in methanogenic environments. Biodegradation 9:463–473
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tang Y-Q, Shigematsu T, Morimura S, Kida K (2007) Effect of dilution rate on the microbial structure of a mesophilic butyrate-degrading methanogenic community during continuous cultivation. Appl Microbiol Biotechnol 75:451–465
Tejayadi S, Cheryan M (1995) Lactic acid from cheese whey permeate. Productivity and economics of a continuous membrane bioreactor. Appl Microbiol Biotechnol 43:242–248
Ueno Y, Haruta S, Ishii M, Igarashi Y (2001) Changes in product formation an bacterial community by dilution rate on carbohydrate fermentation by methanogenic microflora in continuous flow stirred tank reactor. Appl Microbiol Biotechnol 57:65–73
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679
Yu Y, Kim J, Hwang S (2006) Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: population dynamics and community structures. Biotechnol Bioeng 93:424–433
Acknowledgements
This work was financially supported by the Korea Ministry of Knowledge and Economy (MKE) through the Manpower Development Program for Energy and Resources, by the Korea Ministry of Education (MOE) as the Human Resource Development Project for Energy from Waste and Recycling. This work was also partly supported by the Science Foundation Ireland (SFI).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, C., Kim, J., Shin, S.G. et al. Quantitative and qualitative transitions of methanogen community structure during the batch anaerobic digestion of cheese-processing wastewater. Appl Microbiol Biotechnol 87, 1963–1973 (2010). https://doi.org/10.1007/s00253-010-2685-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2685-1