Abstract
The microbial electrolysis cell (MEC) is a promising system for hydrogen production. Still, expensive catalysts such as platinum are needed for efficient hydrogen evolution at the cathode. Recently, the possibility to use a biocathode as an alternative for platinum was shown. The microorganisms involved in hydrogen evolution in such systems are not yet identified. We analyzed the microbial community of a mixed culture biocathode that was enriched in an MEC bioanode. This biocathode produced 1.1 A m−2 and 0.63 m3 H2 m−3 cathode liquid volume per day. The bacterial population consisted of 46% Proteobacteria, 25% Firmicutes, 17% Bacteroidetes, and 12% related to other phyla. The dominant ribotype belonged to the species Desulfovibrio vulgaris. The second major ribotype cluster constituted a novel taxonomic group at the genus level, clustering within uncultured Firmicutes. The third cluster belonged to uncultured Bacteroidetes and grouped in a taxonomic group from which only clones were described before; most of these clones originated from soil samples. The identified novel taxonomic groups developed under environmentally unusual conditions, and this may point to properties that have not been considered before. A pure culture of Desulfovibrio strain G11 inoculated in a cathode of an MEC led to a current development from 0.17 to 0.76 A m−2 in 9 days, and hydrogen gas formation was observed. On the basis of the known characteristics of Desulfovibrio spp., including its ability to produce hydrogen, we propose a mechanism for hydrogen evolution through Desulfovibrio spp. in a biocathode system.
Similar content being viewed by others
Introduction
The high-energy demands of our modern society in combination with the foreseeable depletion of fossil fuels call for the development of sustainable, green forms of energy. Biomass or the organic waste from wastewaters is a source of renewable energy. Recent advances in the use of organic matter for energy production include electricity generation in a microbial fuel cell (MFC) (Logan et al. 2006) and the production of hydrogen in a microbial electrolysis cell (MEC) (Liu et al. 2005; Rozendal et al. 2006; Logan et al. 2008). These kinds of systems are still under development, but they show great potential for green energy production.
Both MFC and MEC usually consist of two compartments containing an anode and a cathode separated by an ion exchange membrane (Rozendal et al. 2007). The two electrodes are connected through an electrical circuit. At the anode, electrochemically active microorganisms are present that consume organic matter and transfer the electrons derived from metabolic processes to the electrode, either by direct or indirect extracellular electron transfer (Ieropoulos 2005; Lovley 2006; Stams et al. 2006; Torres et al. 2009; Lovley and Nevin 2011). An electron acceptor in the cathode liquid enables a current flow from anode to cathode. Typically, oxygen or Fe(III) is used as the electron acceptor in the MFC (Rabaey and Verstraete 2005; Logan and Regan 2006), while in the MEC, protons act as the sole electron acceptor to form hydrogen. For the MEC, a supply of electrical energy is required to make hydrogen gas production possible (Liu et al. 2005; Rozendal et al. 2006).
Acetate is often used as model substrate in MEC systems because it is an end product of fermentation. Theoretically, acetate oxidation yields a potential of −0.29 V (vs. standard hydrogen electrode (SHE), at pH 7, pH2 = 1 bar), while for hydrogen production from protons, a potential of −0.41 V (vs. SHE, at pH 7, pH2 = 1 bar) is required (Liu et al. 2005). Energy is added by applying enough voltage to render an exergonic reaction. Hence, the theoretically applied voltage required for hydrogen gas production in an MEC fed with acetate is 0.12 V. In comparison, for conventional water electrolysis, the theoretically applied voltage needed is 1.2 V at pH 7 (Liu et al. 2005). The lower energy requirement of the MEC makes it an attractive system for hydrogen gas production. In practice, however, a minimum applied voltage of 0.25 V is needed because of several potential losses in the system (Rozendal et al. 2006; Sleutels et al. 2009a, b). The total applied voltage demand in practice is for a great part dependent on the overpotential at the electrodes. The use of a good catalyst can decrease the overpotential significantly (Jeremiasse et al. 2009b). Conventionally, platinum is used as a catalyst for hydrogen gas production (Vetter 1967) and is therefore also applied at MEC cathodes (Rozendal et al. 2006). Because of the high costs and scarcity of platinum, alternative catalysts for hydrogen production are desirable. Microbial cathodes (biocathodes) form an alternative with great prospectives since they are low cost (both electrode material and catalyst) and self-generating. A biocathode can be defined as an electrode from cheap material (e.g., carbon) with a microbial population present at the electrode or in the electrolyte that catalyzes the cathodic reaction. To act as a biocathode in an MEC, microorganisms need to be able to take up electrons from the electrode material and use these electrons to produce hydrogen.
The uptake of electrons from a solid surface or cathode is known from corrosion studies, where metals (e.g., iron) are oxidized by microorganisms that use the electrons from this reaction for metabolic processes (Dinh et al. 2004; Mehanna et al. 2009). Furthermore, in MFCs, biocathodes have been successfully applied to reduce oxygen, fumarate, nitrate, perchlorate, or chlorinated compounds (Huang et al. 2011).
Microorganisms that can produce hydrogen are found in a large variety of environments (Schwartz and Friedrich 2006) and contain hydrogenases that catalyze the reversible reaction 2H+ + 2e− ↔ H2. Purified hydrogenases have been successfully used on carbon electrodes as a catalyst for hydrogen production (Vignais et al. 2001; Lojou and Bianco 2004; Lojou 2011; Vincent et al. 2007). The drawback for these systems is that the enzymes are relatively unstable and lose catalytic activity over time. The use of whole cells can help in maintaining enzyme stability. Immobilization of whole Desulfovibrio vulgaris cells (well known to contain hydrogenases) on an electrode was successful for hydrogen production, and the process was more stable than with enzymes only (Guiral-Brugna et al. 2001; Lojou et al. 2002). For continuous hydrogen production, the challenge is to generate a biocathode with living cells, able to survive and grow.
The microbial uptake of electrons from a cathode for the production of hydrogen in an MEC was shown for the first time by Rozendal et al. (2008). In their study, an MEC half cell with carbon felt electrodes was started up with a biological anode that was initially fed with acetate and hydrogen. Hexacyanoferrate(III) was reduced at the cathode. When stable anodic current was reached, the acetate and hydrogen supply was stopped, and the polarities of anode and cathode were reversed, resulting in a biocathode and chemical anode. The cathode potential was poised at −0.7 V vs. SHE resulting in an average current of 1.1 A m−2 and production of 0.63 m3 H2 m−3 cathode liquid volume per day. A similar setup that was not inoculated served as negative control and produced a current of 0.3 A m−2 and 0.08 m3 H2 m−3 cathode liquid volume per day. In the present study, we describe the microbial population present on the graphite felt cathode using scanning electron microscopy, denaturing gradient gel electrophoresis (DGGE), and cloning and sequencing of 16S ribosomal RNA genes.
Material and methods
Microbial electrolysis cell operation and sample collection
The setup and operation of the microbial electrolysis cell was described previously (Rozendal et al. 2008). From this experiment, samples were collected for the current study. The inoculum of this setup was a mixed microbial community previously enriched and sequentially transferred over a period of 4 years in MFC and MEC anodes amended with acetate. The original MFC anode was inoculated with anaerobic sludge from a paper mill wastewater treatment plant (Eerbeek, the Netherlands), anodic effluent from a molasses-fed MFC, and Geobacter sulfurreducens strain PCA (U. Michaelidou, personal communication). Briefly, the system was started as a two-chamber microbial fuel cell fed with a mixture of acetate and hydrogen (at the bio-electrode) and with a mixed hexacyanoferrate(II) and hexacyanoferrate(III) solution as catholyte (at the chemical counter electrode). The electrode material was graphite felt of 6 mm thickness. After 8 days, the substrate for the bio-electrode was changed to bicarbonate and hydrogen. Following stable anodic current production at a bio-electrode potential of −0.2 V (vs. SHE), at day 11 the hydrogen supply was stopped, and the bio-electrode potential was lowered to −0.7 V. This resulted in consumption of electrons and production of hydrogen at the bio-electrode, now functional as biocathode. The current developed from 0.3 to 1.2 A m−2 projected electrode surface area over a period of 13 days. After this period, bicarbonate addition was stopped. In this layout, i.e., a biocathode with a chemical hexacyanoferrate(II) oxidizing anode, the system was run for over 40 days without addition of any carbon source. Subsequently, the system was disassembled, and samples were collected for our study. Graphite felt electrode material with attached biomass was cut into 7-mm-diameter disks originating from four different locations of the electrode (1) influent site: where medium enters the cathode compartment, (2) middle of the electrode, (3) effluent site: where medium exits the cathode compartment (4) non-flow site: part of electrode which did not have direct contact with the flow path of the medium. Additional liquid samples (2 ml) were taken from the inoculum material (I) and from the effluent (E) at the end of the run. Samples for DNA extraction were stored at −20°C until use.
Strains and cultivation
Desulfovibrio strain G11 (DSM 7057) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). It was cultured with 10 mM lactate as energy and carbon source and 15 mM sulfate as electron acceptor in anaerobic medium with H2 headspace as additional energy source. The medium consisted of (millimolars) MgCl2, 1; CaCl2, 0.7; (NH4)2SO4, 0.4; NH4Cl, 4.7; KH2PO4, 6; and Na2HPO4, 9, supplemented with 1 mL/L trace elements (Zehnder et al. 1980), 1 mL/L selenite/tungstate solution (Widdel and Bak 1992), 2 mL/L vitamins (Wolin et al. 1963), and 0.5 mg/L resazurin. Na2S (1 mM) was added to reduce the media.
Desulfovibrio G11 biocathode
One liter of a Desulfovibrio G11 culture, grown to the end log phase, was centrifuged (8,000 rpm, 15 min), and the pellet was resuspended in 10 mL anoxic phosphate-buffered saline (pH 7.4). The MEC design was as previously described (Jeremiasse et al. 2010) with the exception that both electrodes consisted of 2.5-mm-thick graphite felt, and the membrane was a Ralex cation exchange membrane. The anolyte (hexacyanoferrate(II)) and catholyte (phosphate-buffered medium, as used in Rozendal et al. 2008) recycle speed was 60 mL/min. The cathode was inoculated with 10 mL of cell suspension. After inoculation the cell suspension was pumped to the electrode compartment and left without pumping for 1 h. The phosphate-buffered medium was supplemented with 2 mM of bicarbonate and 0.1 mL/L selenium/tungstate solution; selenium is an essential trace element for some hydrogenases in Desulfovibrio species (Hensgens et al. 1994; Valente et al. 2006). Temperature was controlled at 303 K, and pH was controlled at 7 by dosing 1 M HCl. The cathode potential was manually adjusted to −0.7 V vs. SHE if needed. The current, pH, anode potential, and cathode potential were logged every 5 min (Memograph M, Endress + Hauser, Naarden, the Netherlands). Cathodic current is measured with a negative sign; all current measurements were multiplied by −1 and thereby indicated with a positive sign. When stable current was reached (day 9 after inoculation), a gas sample was taken from the headspace, and H2, CO2 and CH4 were measured using gas chromatography (Varia CP-4900 microGC, TCD detector, MS5 and PPU columns in parallel).
DNA extraction and amplification of 16S rRNA gene
Genomic DNA was extracted from the graphite felt and the liquid samples using the Fast DNA spin kit for soil (Bio101, Vista, CA, USA). Bacterial 16S rRNA genes were amplified with the primers Bact27F and Univ1492R (Lane 1991). PCR settings were initial denaturation for 2 min at 95°C, followed by 25 cycles of 30-s denaturation at 95°C, 40 s annealing at 52°C, and 1.5 min elongation at 72°C. Post-elongation was 5 min at 72°C. The PCR samples were tested on a 1% agarose gel for amount and size of product. Partial bacterial 16S rRNA genes to be used for DGGE analysis were amplified using primers Bact968F (including GC clamp) and 1401R (Nübel et al. 1996). PCR conditions were as above, except that 35 cycles were applied, and an annealing temperature of 56°C was used.
Clone library construction and analysis
For clone library analysis, the electrode sample from the middle of the electrode (2) was used. PCR amplicons of almost complete bacterial 16S rRNA genes were purified using Nucleo Spin Extract II kit (Macherey-Nagel, Düren, Germany) and ligated into pGEM-T easy vector system I (Promega, Madison, WI, USA). After ligation, the vectors were transformed in XL-1 blue competent Escherichia coli cells (Stratagene, Santa Clara, CA, USA) and grown on LB agar containing 100 mg/L ampicillin, 0.1 mM isopropyl-1-thio-β-d-galactopyranoside, and 40 mg/L 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). Fifty-seven white clones were used for further analysis. Clone inserts from lysed E. coli cells (95°C, 10 min) were amplified with primers T7 and SP6 (Promega, Madison, WI, USA) using 35 cycles and an annealing temperature of 55°C. The PCR products were purified using the DNA clean and concentrator-5 kit (Zymo Research, Orange, CA, USA).
Clone inserts were sequenced by the company Baseclear (Leiden, the Netherlands) using primers T7, SP6, and 1100R (Lane 1991). The DNA sequences were checked using Chromas (version 2.32, Technelysium Pty. Ltd.), and contigs were constructed from the partial sequences using DNAbaser (version 2.71.0, Heracle Software, Lilienthal, Germany) resulting in a sequence of (at least) the first 1,250 bp of the 16S rRNA gene. The obtained bacterial 16S rRNA sequences were checked for anomalies using Pintail online software (Ashelford et al. 2005) and compared to the GenBank database using the National Center for Biotechnology Information (NCBI) blastn tool to identify the most closely related sequences. The newly obtained sequences were deposited in the European Molecular Biology Laboratory nucleotide sequence database (accession numbers FR669194-FR669243 and FR675968-FR675974), aligned using the online Silva alignment tool (Pruesse et al. 2007), and merged with the ARB database using ARB software package version 5.1 (Ludwig et al. 2004). A phylogenetic tree was constructed using the ARB Neighbor-Joining Algorithm with bootstrapping (1,000 replicates) and Jukes Cantor correction.
Denaturing gradient gel electrophoresis
The bacterial communities populating the graphite electrode on four different spots of the electrode, as well as inoculum and effluent samples, were analyzed with DGGE. Amplicons were run on an 8% polyacrylamide gel containing a formamide and urea denaturant gradient of 30–60%, similar as described by Martín et al. (2007). Gels were run for 16 h at 60°C and stained with silver nitrate (Sanguinetti et al. 1994). In addition, all amplicons of the clones were run on DGGE to evaluate their positions on the gel in comparison to the band migration behavior of the total bacterial communities. The bands that were not identified from the clones were excised from the gel. This material was incubated in Tris–EDTA buffer (10 mM Tris and 1 mM EDTA, pH 8) at 4°C for 2 days, and the extract was used for re-amplification with DGGE primers. The PCR product was checked for purity on DGGE, purified and sequenced with primers 968F by Baseclear (Leiden, the Netherlands). The obtained sequences were checked in Chromas and the most closely related relatives identified using the NCBI blastn search tool.
Results
Bacterial community on the electrode
During operation the MEC produced 0.63 m3 H2 m−3 cathode liquid volume per day or 0.63 × 10−3 m3 H2 m−2 electrode surface per day at an applied voltage of −0.7 V vs. SHE, and scanning electron microscopic imaging showed that after more than 80 days of operation, microorganisms were present and attached to the electrode felt fibers (Rozendal et al. 2008). Analysis of the dominant members of the bacterial population on the graphite felt electrode by DGGE showed a complex pattern which did not differ between the different locations on the electrode (Fig. 1), but there was a large difference between inoculation material, effluent material, and electrode material. A large number of bands with different intensity were present, presumably reflecting the difference in abundance of different bacterial ribotypes. Subsequent cloning and sequencing of the bacterial community were done with the sample obtained from the middle of the electrode.
DGGE profile of the total bacterial community on the biocathode. a Different locations and samples. I inoculation material, 1 influent site of electrode compartment, 2 middle of electrode, 3 effluent site of the electrode compartment, 4 non-flow site of the electrode, E effluent liquid. b Identification of the bands from sample 2. Identity of the bands is indicated with the matching clone named in brackets; B112Dh (FR669244) was identified by PCR amplification of the band cut from the DGGE profile. The indicated percentages are percentage identity of the found sequence with the indicated closest relatives found in GenBank
16S rRNA gene distribution among different phylogenetic groups
The sequences from the 16S rRNA gene clone library (total of 57 clones) were related to seven different phyla (Table 1). The majority (88%) of the clones clustered within three phyla, the Proteobacteria (26 clones, 46%, FR669218-FR669243), the Firmicutes (14 clones, 25%, FR669204-FR669217), and the Bacteriodetes (10 clones, 17%, FR669194-FR669203). The other 12% of the clones were phylogenetically related to Tenericutes (one clone, 2%, FR675972), Spirochaetes (one clone, 2%, FR675968), Chlorobi (one clone, 2%, FR675969), Actinobacteria (two clones, 3%, FR675973 and FR675974), and unclassified bacteria (two clones, 3%, FR675970 and FR675971).
The phylogenetic relation of the clones from our study, including several related uncultured and cultured species from the GenBank database, is shown in Fig. 2. Within the Proteobacteria, 81% (21 clones) belonged to the subclass of the Deltaproteobacteria of which 90% (19 clones) of the clones clustered with the genus Desulfovibrio (Fig. 2a). Moreover, 67% (14 clones) showed more than 98% identity to the species D. vulgaris Hildenborough, an anaerobic bacterium that is able to use hydrogen, organic acids, or alcohols as electron donor and sulfate as electron acceptor (Postgate and Campbell 1966). D. vulgaris is also known to be able to produce hydrogen (Carepo et al. 2002). The remaining 10% of Deltaproteobacteria (two clones) showed identical 16S rRNA gene sequences to G. sulfurreducens strain PCA, a bacterium which uses hydrogen or acetate as electron donor to reduce metals (Caccavo et al. 1994). G. sulfurreducens is able to use the electrode of an MFC as electron acceptor (Bond and Lovley 2003) but also as electron donor for fumarate reduction (Gregory et al. 2004; Dumas et al. 2008) and hydrogen production (Geelhoed and Stams 2011). The other 19% (four clones) of Proteobacteria belonged to the Alpha- (one clone), Beta- (two clones), and Gammaproteobacteria (one clone) and were most closely related to, respectively, Mesorhizobium amorphae, Pelomonas saccharophila, Azonexus caeni, and Dokdonellala koreensis (Fig. 2a).
Phylogenetic neighbor-joining bootstrap (1,000 replicates) consensus tree of the three major phyla detected on the biocathode. a Proteobacteria (accession numbers: FR669218-FR669243), b Firmicutes (accession numbers: FR669204-FR669217), c Bacteroidetes (accession numbers: FR669194-FR669203). The archaeon Thermofilum pendense is used as outgroup. Clones from our study are indicated in bold, and the rest represent 16S rDNA sequences imported from the GenBank database. The sequences in the tree are at least 1,250 bp long. The scale bar indicates the distance of 0.05 (or 5%) sequence identity
All 14 clones that clustered in the phylum of Firmicutes belonged to the Clostridia class. Within the Clostridia, one prominent group of nine clones (64% of the total Firmicutes) belonged to the family of unclassified Clostridiales (Fig. 2b). The clones in this group showed more than 98% identity to each other but did not belong to any cultured or uncultured genus listed in the GenBank database (using 95% identity in the 16S rRNA gene as the genus delineation value (Rosselló-Mora and Amann 2001)). The most similar GenBank sequence showed only 90% identity to this cluster of uncultured Firmicutes and was obtained from a dechlorinating flow column (FM178833) (Behrens et al. 2008). The most closely related cultured bacterium was Desulfitobacterium hafniense, with 88% 16S rRNA gene sequence identity. The other clones within the Firmicutes were all related to anaerobic species such as Clostridium viride, Clostridium cylindrosporum, or uncultured species found in anaerobic environments such as dechlorinating enrichment cultures (EF64459), anaerobic digesters (CU918499 and CR933290), mouse cecum (EU457310), a trichlorobenzene transforming culture (AJ009499), and a denitrifying community inoculated from rice paddy soil (AB486915).
Within the phylum of Bacteroidetes, seven clones (70%) belonged to the class of Bacteroidia. Within this class, there were five clones that clustered together as unclassified Bacteroidetes with more than 99% identity to each other. These five clones did not belong to any cultured genus. In the GenBank database, several sequences were present that clustered in the same taxonomic group (more than 95% identity). These sequences were derived from the following sources: chloraminated drinking water distribution system (EU808333), chromium-contaminated soil (EU037360), and high-carbohydrate and high-pH sludge (FJ5234992) (Fig. 2c). The closest related cultured microorganism was Rikenella microfusus with 92% identity of the 16S rRNA gene. R. microfusus is a fermentative bacterium isolated from feces of a Japanese quail (Kaneuchi and Mitsuoka 1978). The other clones within the class of Bacteroidia were related to uncultured species from nitrobenzene-polluted river water (EF590019) and a bovine serum albumin digester (AB175369). The other clones within the Bacteroidetes phylum belonged to the class of uncultured Sphingobacteria (two clones, 20%) and Sediminibacteria (one clone, 10%) and were most closely related to uncultured bacteria from thermophilic anaerobic sludge fed with methanol (AY526509) and Sediminibacterium salmoneum, an aerobic bacterium isolated from sediment of the Guanting reservoir in Beijing, China.
Identification of cloned 16S rDNA in the bacterial DGGE profile
Comparison of the sequencing data and the DGGE profiles of the clones and the total bacterial community showed that the most dominant band represented D. vulgaris (clone 1B). The other identified bands represented other Desulfovibrio sp. (B112Dh, FR669244), G. sulfurreducens (clone 6C), uncultured Bacteroidetes (clone 3E and clone 5D), an uncultured Firmicutes sp. (clone 12A), and an uncultured Actinobacterium (clone 5H) (Fig. 1).
A pure culture of Desulfovibrio G11 in an MEC
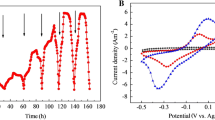
After inoculation of Desulfovibrio G11 in the MEC cathode, the current increased from 1.7 (0.17 A m−2) to 7.6 mA (0.76 A m−2) over a period of 9 days (Fig. 3). During this period, it was visible that gas accumulated in the system. Analysis of the gas phase on day 9 showed the presence of hydrogen gas, whereas no methane or carbon dioxide was detected.
Current production over time after inoculation of the cathode with pure culture of Desulfovibrio G11. The current increased from 0.17 to 0.76 A m−2 in 9 days. Arrow 1 indicates the time of inoculation; arrows 2, 3, and 4 indicate an adjustment of the cathode potential to −0.7 V vs. SHE; and arrow 5 indicates the time of gas sampling
Discussion
In previous research, hydrogen production in an MEC with a biocathode has been shown (Rozendal et al. 2008; Jeremiasse et al. 2009a). Our research gives the first description of a microbial community of a hydrogen-producing biocathode in an MEC. The results showed high bacterial 16S rRNA gene diversity, with the dominant species belonging to the genus Desulfovibrio. Two other predominant clusters were found that were related to uncultured Firmicutes and uncultured Bacteroidetes. In addition to being the dominant ribotype in the MEC biocathode, progression of current was shown after inoculation of an MEC cathode with pure cultures of Desulfovibrio G11.
The dominance of Desulfovibrio spp. in the biocathode can be reasoned because Desulfovibrio species are well known for their ability to produce and consume hydrogen gas (Carepo et al. 2002). Coating of an electrode with immobilized D. vulgaris cells has been reported to catalyze the evolution of hydrogen at a cathode (Lojou et al. 2002). However, this catalytic effect occurred only in the presence of the electron shuttle methyl viologen. The application of living Desulfovibrio as hydrogen catalyst at a cathode without an added mediator, as in our system, was not shown before. This is not only fundamentally, but also practically, of great interest because it will allow low-cost and self-maintaining cathode systems for hydrogen production.
The second major group of bacteria found in this study, the uncultured Firmicutes, does not belong to any earlier described genus. Apparently, the conditions in the MEC created an environment in which bacteria belonging to a new taxonomic group were able to develop predominantly. Moreover, it is interesting to note that the closest related genus is Desulfitobacterium, of which at least one member, D. hafniense strain DCB2, was electrochemically active in an anode from an MFC (Milliken and May 2007). Furthermore, Desulfitobacterium spp. were found as the dominant population in a mixed culture that was producing hydrogen gas in a dechlorinating cathode system (Aulenta et al. 2008). Similar to the experiment with immobilized D. vulgaris, no hydrogen was produced in the absence of methyl viologen as a mediator.
The third major group of bacteria in the MEC biocathode belonged to the uncultured Bacteroidetes and also constitutes a novel group without cultured relatives at genus level (92% identity). Members of the Bacteroidetes phylum are found in a large variety of environments such as soil, sediments, human and animal gut, and seawater. The group of uncultured Bacteroidetes clones in our study was most closely related to clones from various environmental samples that presumably all originated from anaerobic sources. The closest related cultured bacterium, R. microfusus, grows fermentatively on carbohydrates (Kaneuchi and Mitsuoka 1978), but no other information about its metabolism is available.
The principle of electron uptake from a solid surface has been shown before, but the mechanisms are poorly understood. The reverse process of electron transfer to an anode has been studied in more detail, and those studies provided information on which mechanisms are possible (Rabaey et al. 2004; Lovley 2008; Nevin et al. 2009). Extracellular electron transfer can take place indirectly using electron shuttles such as methyl viologen, humic acid, sulfide, cysteine, riboflavin, phenazine, and quinones (Stams et al. 2006; Logan 2009). Membrane-associated proteins such as cytochromes and cell appendages or nanowires have been suggested to be involved in direct electron transfer (Kim 2002; Mehta et al. 2005; Reguera et al. 2005). For the most extensively studied species G. sulfurreducens, expression and deletion studies have shown that direct extracellular electron transfer to an electrode involves multicopper proteins (Holmes et al. 2008), several c-type cytochromes, (Holmes et al. 2006) and pillin structures which most likely are involved in the physical association with the electrode (Nevin et al. 2009). Furthermore, G. sulfurreducens can change from electron donating to electron uptake for hydrogen production after reversing the potential from anodic to cathodic (Geelhoed and Stams 2011). For extracellular electron uptake from an electrode, several mechanisms have been suggested (Geelhoed et al. 2010; Rosenbaum et al. 2011). Recent findings suggest that G. sulfurreducens uses different cytochromes in the pathways for electron donating than for electron uptake (Strycharz et al. 2011). These authors suggest that this might reflect the optimal potential at which specific proteins can accept or donate electrons. With our findings that Desulfovibrio spp. are dominant microorganisms at the cathode together with our findings that Desulfovibrio G11 is electrochemically active at the cathode, the possible mechanisms of electron transfer and hydrogen production for this species can be inferred, as discussed below.
The genomes of Desulfovibrio species show several c-type cytochromes and multicopper proteins with homology to the proteins involved in electron donation in Geobacter species (NCBI search). Similar to the pillin structures in Geobacter spp., D. vulgaris flagellar appendages (genes flgC, flgB, and flgL) have been associated with physical association during syntrophic growth (Walker et al. 2009) and might also be involved in adherence to electrodes. These similarities suggest that the mechanism of extracellular electron transfer by Desulfovibrio spp. could be similar to previously described mechanisms of electron transfer involving extracellular appendage (pili or flagella)-like structures, cytochromes, or shuttle compounds. The electron transfer from an electrode to the microorganisms can possibly take place by reversed reaction of those previously described mechanisms. More research is needed to understand how electron transfer in cathode systems takes place.
Hydrogen production from protons is energetically costly. For microbial hydrogen production, energy needs to be added in the form of an electron donor with high energy (e.g., glucose or light) or in the MEC biocathode by the applied voltage. A putative mechanism for the conservation of energy from hydrogen production at the cathode may be comparable to hydrogen production from formate in methanogenic co-cultures (Dolfing et al. 2008; Stams and Plugge 2009). Energy gain and growth from production of hydrogen have been shown for Desulfovibrio G11 grown on formate in coculture with Methanobrevibacter arboriphilus AZ (Dolfing et al. 2008). Conservation of energy by Desulfovibrio spp. was proposed to involve an energy-conserving hydrogenase or a hydrogenase present at the cytoplasmic side of the membrane. In the genome of D. vulgaris, genes coding for both types of hydrogenases are present. The release of protons from formate by a formate dehydrogenase located at the periplasmic side of the membrane combined with proton consumption at the cytoplasmic side results in the generation of a proton gradient over the membrane that can be utilized by a membrane-bound ATPase. It has been suggested that in a similar way energy, could be gained from the production of hydrogen from electrons derived from a cathode (Geelhoed et al. 2010). Our findings that Desulfovibrio species dominate the microbial community of the MEC support the idea that the mechanism of electron transfer from an electrode to the bacterium can take place like suggested before by Dolfing et al. (2008) and Geelhoed et al. (2010). Comparing MEC experiments to syntrophic growth, Geelhoed et al. (2010) calculated that the energy applied to MEC systems is enough to allow energy conservation and growth. However, in those calculations, no energy losses in the system were taken into account. The actual energy available at the cathode can be estimated from the potential of the cathode (−0.7 V vs. SHE) minus the energy needed to form hydrogen (−0.41 V vs. SHE) which gives the maximum theoretical energy available for the microorganisms (−0.29 V). The cathode losses, expressed as the concentration overpotential, can be calculated as described by Jeremiasse et al. (2009b). Under the conditions prevailing in the biocathode system studied here (Rozendal et al. 2008), using a pKa2 of 7.21 for phosphate buffer, the concentration overpotential can be estimated at −0.019 V. Hence, the actual energy available for the microorganisms is −0.29 + 0.019 = −0.27 V or −52 kJ per mole H2 produced (at pH 7 and pH2 = 1 bar). In comparison, the Gibbs free energy change associated with conversion of formate to hydrogen and carbon dioxide is −17 to −19 kJ per mole H2 (Dolfing et al. 2008). This shows that for the studied biocathode system, even if the overpotential is taken into account, there is sufficient energy available for the microorganisms to grow. The energetic limits for microbial hydrogen production and growth in an MEC biocathode still need to be explored.
Our findings that the dominant microorganism in the MEC biocathode is a Desulfovibrio sp., together with the knowledge about the hydrogen metabolism and potential for exocellular electron transfer of Desulfovibrio spp., give very strong indications that they are actively involved in the hydrogen production at the biocathode of the MEC. Since Desulfovibrio spp. are also able to consume hydrogen, it can be reasoned that the microbial community on the electrode developed during the anodic phase, in which acetate and hydrogen were the substrates. However, after switching the polarity, the production of hydrogen gas commenced only after several days, suggesting that microbial adaptation and possibly growth were necessary to start hydrogen production at the cathode. In addition, the potential electroactivity of Desulfovibrio in a cathode was supported by the observed current production and hydrogen production after inoculation of an MEC cathode with Desulfovibrio G11. Besides Desulfovibrio, two novel and abundantly present groups of bacteria were present. These bacteria need to be characterized further before their role in an MEC can be inferred.
References
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16rRNA sequences records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 12:7724–7736
Aulenta F, Canosa A, Majone M, Panero S, Reale P, Rossetti S (2008) Trichloroethene dechlorination and H2 evolution are alternative biological pathways of electric charge utilization by a dechlorinating culture in a bioelectrochemical system. Environ Sci Technol 42:6185–6190
Behrens S, Azizian MF, McMurdie PJ, Sabalowsky A, Dolan ME, Semprini L, Spormann AM (2008) Monitoring abundance and expression of “Dehalococcoides” species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Appl Environ Microbiol 74:5695–5703
Bond DR, Lovley DR (2003) Electricity production by Geobacter sulfurreducens attached to electrodes. Appl Environ Microbiol 69:1548–1555
Caccavo F, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ (1994) Geobacter sulfurreducens sp. nov, a hydrogen-oxidizing and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol 60:3752–3759
Carepo M, Baptista JF, Pamplona A, Fauque G, Moura JJG, Reis MAM (2002) Hydrogen metabolism in Desulfovibrio desulfuricans strain New Jersey (NCIMB 8313)—comparative study with D. vulgaris and D. gigas species. Anaerobe 8:325–332
Dinh HT, Kuever J, Mussmann M, Hassel AW, Stratmann M, Widdel F (2004) Iron corrosion by novel anaerobic microorganisms. Nature 427:829–832
Dolfing J, Jiang B, Henstra AM, Stams AJM, Plugge CM (2008) Syntrophic growth on formate: a new microbial niche in anoxic environments. Appl Environ Microbiol 74:6126–6131
Dumas C, Basseguy R, Bergel A (2008) Microbial electrolysis with Geobacter sulfurreducens biofilm on stainless steel cathode. Electrochim Acta 53:2494–2500
Geelhoed JS, Stams AJM (2011) Electricity-assisted biological hydrogen production from acetate by Geobacter sulfurreducens. Environ Sci Technol 45:815–820
Geelhoed JS, Hamelers HVM, Stams AJM (2010) Electricity-mediated biological hydrogen production. Curr Opin Microbiol 13:307–315
Gregory KB, Bond DR, Lovley DR (2004) Graphite electrode as electron donors for anaerobic respiration. Environ Microbiol 6:596–604
Guiral-Brugna M, Giudici-Orticoni MT, Bruschi M, Bianco P (2001) Electrocatalysis of the hydrogen production by [Fe] hydrogenase from Desulfovibrio vulgaris Hildenborough. J Electroanal Chem 510:136–143
Hensgens CMH, Nienhuis-Kuiper ME, Hansen TA (1994) Effect of tungstate on the growth of Desulfovibrio gigas NCIMB 9332 and other sulfate-reducing bacteria with ethanol as a substrate. Arch Microbiol 162:143–147
Holmes DE, Chaudhuri SK, Nevin KP, Mehta T, Methé BA, Liu A, Ward JE, Woodard TL, Webster J, Lovley DR (2006) Microarray and genetic analysis of electron transfer to electrodes in Geobacter sulfurreducens. Environ Microbiol 8:1805–1815
Holmes DE, Mester T, O'Neil RA, Perpetua LA, Larrahondo MJ, Glaven R, Sharma ML, Ward JE, Nevin KP, Lovley DR (2008) Genes for two multicopper proteins required for Fe(III) oxide reduction in Geobacter sulfurreducens have different expression patterns both in the subsurface and on energy-harvesting electrodes. Microbiology 154:1422–1435
Huang L, Regan JM, Quan X (2011) Electron transfer mechanisms, new applications, and performance of biocathode microbial fuel cells. Biores Technol 102:316–323
Ieropoulos IA (2005) Comparative study of three types of microbial fuel cell. Enzyme Microb Tech 37:238–245
Jeremiasse AW, Hamelers HVM, Buisman CJN (2009a) Microbial electrolysis cell with a microbial biocathode. Bioelectrochem 78:39–43
Jeremiasse AW, Hamelers HVM, Kleijn JM, Buisman CJN (2009b) Use of biocompatible buffers to reduce the concentration overpotential for hydrogen evolution. Environ Sci Technol 43:6882–6887
Jeremiasse AW, Hamelers HVM, Saakes M, Buisman CJN (2010) Ni foam cathode enables high volumetric H2 production in a microbial electrolysis cell. Int J Hydrogen Energy 35:12716–12723
Kaneuchi C, Mitsuoka T (1978) Bacteroides microfusus, a new species from intestines of calves, chickens, and Japanese quails. Int J Syst Bacteriol 28:478–481
Kim HJ (2002) A mediator-less microbial fuel cell using a metal reducing bacterium, Shewanella putrefaciens. Enzyme Microb Tech 30:145–152
Lane DL (1991) 16S/23S rRNA sequencing. In: Stackebrandt ER, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Liu H, Grot S, Logan BE (2005) Electrochemically assisted microbial production of hydrogen from acetate. Environ Sci Technol 39:4317–4320
Logan BE (2009) Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol 7:375–381
Logan BE, Regan JM (2006) Electricity-producing bacterial communities in microbial fuel cells. Trends Microbiol 14:512–518
Logan BE, Hamelers HVM, Rozendal RA, Schröder U, Keller J, Freguia S, Aelterman P, Verstraete W, Rabaey K (2006) Microbial fuel cells: methodology and technology. Environ Sci Technol 40:5181–5192
Logan BE, Call D, Cheng S, Hamelers HVM, Sleutels THJA, Jeremiasse AW, Rozendal RA (2008) Microbial electrolysis cells for high yield hydrogen gas production from organic matter. Environ Sci Technol 42:8630–8640
Lojou E (2011) Hydrogenases as catalysts for fuel cells: strategies for efficient immobilization at electrode interfaces. Electrocim Acta. doi:10.1016/j.electacta.2011.03.002
Lojou EB, Bianco P (2004) Electrocatalytic reactions at hydrogenase-modified electrodes and their applications to biosensors: from the isolated enzymes to the whole cells. Electroanalysis 16:1093–1100
Lojou E, Durand MC, Dolla A, Bianco P (2002) Hydrogenase activity control at Desulfovibrio vulgaris cell-coated carbon electrodes: biochemical and chemical factors influencing the mediated bioelectrocatalysis. Electroanalysis 14:913–922
Lovley DR (2006) Bug juice: harvesting electricity with microorganisms. Nat Rev Microbiol 4:497–508
Lovley DR (2008) The microbe electric: conversion of organic matter to electricity. Curr Opin Biotech 19:564–571
Lovley DR, Nevin KP (2011) A shift in the current: new applications and concepts for microbe-electrode electron exchange. Curr Opin Biotech 22:1–8
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar BA, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Martín R, Heilig GHJ, Zoetendal EG, Smidt H, Rodriguez JM (2007) Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol 103:2638–2644
Mehanna M, Basseguy R, Delia ML, Bergel A (2009) Role of direct microbial electron transfer in corrosion of steels. Electrochem Commun 11:568–571
Mehta T, Coppi MV, Childers SE, Lovley DR (2005) Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol 71:8634–8641
Milliken CE, May HD (2007) Sustained generation of electricity by the spore-forming, Gram-positive, Desulfitobacterium hafniense strain DCB2. Appl Microbiol Biotechnol 73:1180–1189
Nevin KP, Kim BC, Glaven RH, Johnson JP, Woodard TL, Methe BA, DiDonato RJ, Covalla SF, Franks AE, Liu A, Lovley DR (2009) Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4:e5628
Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, Ludwig W, Backhaus H (1996) Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178:5636–5643
Postgate JR, Campbell LL (1966) Classification of Desulfovibrio species, the nonsporulating sulfate-reducing bacteria. Bacteriol Rev 30:732–738
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rabaey K, Verstraete W (2005) Microbial fuel cells: novel biotechnology for energy generation. Trends Biotechnol 23:291–298
Rabaey K, Boon N, Siciliano SD, Verhaege M, Verstraete W (2004) Biofuel cells select for microbial consortia that self-mediate electron transfer. Appl Environ Microbiol 70:5373–5382
Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR (2005) Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101
Rosenbaum M, Aulenta F, Villano M, Angenent LT (2011) Cathodes as electron donor for microbial metabolism: which extracellular electron transfer mechanisms are involved? Biores Technol 102:324–333
Rosselló-Mora R, Amann R (2001) The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67
Rozendal RA, Hamelers HVM, Euverink GJW, Metz SJ, Buisman CJN (2006) Principle and perspectives of hydrogen production through biocatalyzed electrolysis. Int J Hydrogen Energy 31:1632–1640
Rozendal RA, Hamelers HVM, Molenkamp RJ, Buisman CJN (2007) Performance of single chamber biocatalyzed electrolysis with different types of ion exchange membranes. Water Res 41:1984–1994
Rozendal RA, Jeremiasse AW, Hamelers HVM, Buisman CJN (2008) Hydrogen production with a microbial biocathode. Environ Sci Technol 42:629–634
Sanguinetti CJ, Neto ED, Simpson AJG (1994) Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17:914–921
Schwartz E, Friedrich B (2006) The H2-metabolizing prokaryotes. In: Lengeler JW, Drews G, Schlegel HG (eds) Prokaryotes. Thieme, Stuttgart, pp 497–563
Sleutels THJA, Hamelers HVM, Rozendal RA, Buisman CJN (2009a) Ion transport resistance in microbial electrolysis cells with anion and cation exchange membranes. Int J Hydrogen Energy 34:3612–3620
Sleutels THJA, Lodder R, Hamelers HVM, Buisman CJN (2009b) Improved performance of porous bio-anodes in microbial electrolysis cells by enhancing mass and charge transport. Int J Hydrogen Energy 34:9655–9661
Stams AJM, Plugge CM (2009) Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7:568–577
Stams AJM, de Bok FA, Plugge CM, van Eekert MH, Dolfing J, Schraa G (2006) Exocellular electron transfer in anaerobic microbial communities. Environ Microbiol 8:371–382
Strycharz SM, Glaven RH, Coppi MV, Gannon SM, Perpetua LA, Liu A, Nevin KP, Lovley DR (2011) Gene expression and deletion analysis of mechanisms for electron transfer from electrodes to Geobacter sulfurreducens. Bioelectrochem 80:142–150
Torres CI, Marcus AK, Lee HS, Parameswaran P, Krajmalnik-Brown R, Rittmann BE (2009) A kinetic perspective on extracellular electron transfer by anode-respiring bacteria. FEMS Microbiol Rev 34:3–17
Valente FAM, Almeida CC, Pacheco I, Carita J, Saraiva LM, Pereira IAC (2006) Selenium is involved in regulation of periplasmic hydrogenase gene expression in Desulfovibrio vulgaris Hildenborough. J Bacteriol 188:3228–3235
Vetter KJ (1967) Electrochemical kinetics: theoretical and experimental aspects. Academic, New York
Vignais PM, Billoud B, Meyer J (2001) Classification and phylogeny of hydrogenases. FEMS Microbiol Rev 25:455–501
Vincent KA, Parkin A, Armstrong FA (2007) Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem Rev 107:4366–4413
Walker CB, He ZL, Yang ZK, Ringbauer JA, He Q, Zhou JH, Voordouw G, Wall JD, Arkin AP, Hazen TC, Stolyar S, Stahl DA (2009) The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol 191:5793–5801
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes. Springer, New York, pp 3352–3378
Wolin EA, Wolin MJ, Wolfe RS (1963) Formation of methane by bacterial extract. J Biol Chem 238:2882–2886
Zehnder AJB, Huser BA, Brock TD, Wuhrmann K (1980) Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol 124:1–11
Acknowledgments
We thank René Rozendal and Adriaan Jeremiasse for supplying samples from the biocathode. Financial support was obtained from Wetsus and the Chemical Sciences Division of the Netherlands Organisation for Scientific Research, grant CW-TOP 700.55.343. Wetsus is funded by the Dutch Ministry of Economic Affairs, the European Union Regional Development Fund, the province of Friesland, the City of Leeuwarden, and the EZ/Kompas program of the “Samenwerkingsverband Noord-Nederland.” The fruitful discussions with Caroline Plugge, Adriaan Jeremiasse, Urania Michaelidou, and with the members of the Wetsus BioEnergy theme are highly appreciated.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Croese, E., Pereira, M.A., Euverink, GJ.W. et al. Analysis of the microbial community of the biocathode of a hydrogen-producing microbial electrolysis cell. Appl Microbiol Biotechnol 92, 1083–1093 (2011). https://doi.org/10.1007/s00253-011-3583-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3583-x