Abstract
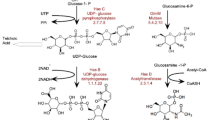
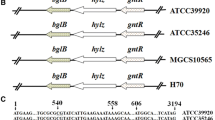
Xylose is described as a component of bacterial exopolysaccharides in only a limited number of bacterial strains. A bacterial strain, Paenibacillus elgii, B69 was shown to be efficient in producing a xylose-containing exopolysaccharide. Sequence analysis was performed to identify the genes encoding the uridine diphosphate (UDP)-glucuronic acid decarboxylase required for the synthesis of UDP-xylose, the precursor of the exopolysaccharide. Two sequences, designated as Peuxs1 and Peuxs2, were found as the candidate genes for such enzymes. The activities of the UDP-glucuronic acid decarboxylases were proven by heterologous expression and real-time nuclear magnetic resonance analysis. The intracellular activity and effect of these genes on the synthesis of exopolysaccharide were further investigated by developing a thymidylate synthase based knockout system. This system was used to substitute the conventional antibiotic resistance gene system in P. elgii, a natural multi-antibiotic resistant strain. Results of intracellular nucleotide sugar analysis showed that the intracellular UDP-xylose and UDP-glucuronic acid levels were affected in Peuxs1 or Peuxs2 knockout strains. The knockout of either Peuxs1 or Peuxs2 reduced the polysaccharide production and changed the monosaccharide ratio. No polysaccharide was found in the Peuxs1/Peuxs2 double knockout strain. Our results show that P. elgii can be efficient in forming UDP-xylose, which is then used for the synthesis of xylose-containing exopolysaccharide.




Similar content being viewed by others
References
Ankel H, Feingold DS (1966) Biosynthesis of uridine diphosphate d-xylose. II. Uridine diphosphate d-glucuronate carboxy-lyase of Cryptococcus laurentii. Biochemistry 5(1):182–189
Bar-Peled M, Griffith CL, Doering TL (2001) Functional cloning and characterization of a UDP- glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc Natl Acad Sci 98(21):12003–12008. doi:10.1073/pnas.211229198
Belfort M, Maley G, Pedersen-Lane J, Maley F (1983) Primary structure of the Escherichia coli thyA gene and its thymidylate synthase product. Proc Natl Acad Sci 80(16):4914–4918
Bindschedler L, Wheatley E, Gay E, Cole J, Cottage A, Bolwell GP (2005) Characterisation and expression of the pathway from UDP-glucose to UDP-xylose in differentiating tobacco tissue. Plant Mol Biol 57(2):285–301. doi:10.1007/s11103-004-7795-7
Bloemberg GV, Lugtenberg BJJ (2001) Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol 4(4):343–350. doi:10.1016/S1369-5266(00)00183-7
Breazeale SD, Ribeiro AA, Raetz CRH (2002) Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid a species modified with 4-amino-4-deoxy-L-arabinose. J Biol Chem 277(4):2886–2896. doi:10.1074/jbc.M109377200
Bystrova OV, Knirel YA, Lindner B, Kocharova NA, Kondakova AN, Zähringer U, Pier GB (2005) Structures of the core oligosaccharide and O‐units in the R‐and SR‐type lipopolysaccharides of reference strains of Pseudomonas aeruginosa O‐serogroups. FEMS Immunol Med Microbiol 46(1):85–99
Coyne MJ, Fletcher CM, Reinap B, Comstock LE (2011) UDP-glucuronic acid decarboxylases of Bacteroides fragilis and their prevalence in bacteria. J Bacteriol 193(19):5252–5259. doi:10.1128/jb.05337-11
Ding R, Li Y, Qian C, Wu X (2011) Draft genome sequence of Paenibacillus elgii B69, a strain with broad antimicrobial activity. J Bacteriol 193(17):4537
Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci 76(4):1648–1652
Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE (2007) Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci 104(7):2413–2418
Götting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K (2000) Molecular cloning and expression of human UDP-D-xylose:proteoglycan core protein β-D-Xylosyltransferase and its first isoform XT-II. J Mol Biol 304(4):517–528. doi:10.1006/jmbi.2000.4261
Gu X, Glushka J, Yin Y, Xu Y, Denny T, Smith J, Jiang Y, Bar-Peled M (2010) Identification of a bifunctional UDP-4-keto-pentose/UDP-xylose synthase in the plant pathogenic bacterium Ralstonia solanacearum strain GMI1000, a distinct member of the 4, 6-dehydratase and decarboxylase family. J Biol Chem 285(12):9030–9040
Gu X, Lee SG, Bar-Peled M (2011) Biosynthesis of UDP-xylose and UDP-arabinose in Sinorhizobium meliloti 1021: first characterization of a bacterial UDP-xylose synthase, and UDP-xylose 4-epimerase. Microbiology 157(1):260–269
Guyett P, Glushka J, Gu X, Bar-Peled M (2009) Real-time NMR monitoring of intermediates and labile products of the bifunctional enzyme UDP-apiose/UDP-xylose synthase. Carbohydr Res 344(9):1072–1078. doi:10.1016/j.carres.2009.03.026
Harper AD, Bar-Peled M (2002) Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, uxs, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol 130(4):2188–2198. doi:10.1104/pp. 009654
Hong J-H, Jung H (2014) Antioxidant and antitumor activities of β-glucan-rich exopolysaccharides with different molecular weight from Paenibacillus polymyxa JB115. J Korean Soc Appl Biol Chem 57(1):105–112. doi:10.1007/s13765-013-4252-9
Hung R-J, Chien H-S, Lin R-Z, Lin C-T, Vatsyayan J, Peng H-L, Chang H-Y (2007) Comparative analysis of two UDP-glucose dehydrogenases in Pseudomonas aeruginosa PAO1. J Biol Chem 282(24):17738–17748. doi:10.1074/jbc.M701824200
Jacobson ES, Payne WR (1982) UDP-glucuronate decarboxylase and synthesis of capsular polysaccharide in Cryptococcus neoformans. J Bacteriol 152(2):932–934
Jones TM, Albersheim P (1972) A gas chromatographic method for the determination of aldose and uronic acid constituents of plant cell wall polysaccharides. Plant Physiol 49(6):926–936
Kochanowski N, Blanchard F, Cacan R, Chirat F, Guedon E, Marc A, Goergen JL (2006) Intracellular nucleotide and nucleotide sugar contents of cultured CHO cells determined by a fast, sensitive, and high-resolution ion-pair RP-HPLC. Anal Biochem 348(2):243–251. doi:10.1016/j.ab.2005.10.027
Kuhn J, Götting C, Schnölzer M, Kempf T, Brinkmann T, Kleesiek K (2001) First isolation of human UDP-D-Xylose: proteoglycan core protein β-D-xylosyltransferase secreted from cultured JAR Choriocarcinoma cells. J Biol Chem 276(7):4940–4947. doi:10.1074/jbc.M005111200
Lenz O, Friedrich B (1998) A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc Natl Acad Sci 95(21):12474–12479. doi:10.1073/pnas.95.21.12474
Li J, Kisara K, Danielsson S, Lindström ME, Gellerstedt G (2007) An improved methodology for the quantification of uronic acid units in xylans and other polysaccharides. Carbohydr Res 342(11):1442–1449. doi:10.1016/j.carres.2007.03.031
Li O, Lu C, Liu A, Zhu L, Wang P-M, Qian C-D, Jiang X-H, Wu X-C (2013) Optimization and characterization of polysaccharide-based bioflocculant produced by Paenibacillus elgii B69 and its application in wastewater treatment. Bioresour Technol 134:87–93. doi:10.1016/j.biortech.2013.02.013
Liu J, Luo J, Ye H, Sun Y, Lu Z, Zeng X (2010) In vitro and in vivo antioxidant activity of exopolysaccharides from endophytic bacterium Paenibacillus polymyxa EJS-3. Carbohydr Polym 82(4):1278–1283
Mishra A, Jha B (2013) Microbial exopolysaccharides. In: Rosenberg E, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes: applied bacteriology and biotechnology, 4th edn. Springer, Berlin, pp 179–192
Mohnen D (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11(3):266–277. doi:10.1016/j.pbi.2008.03.006
Mokaddem H, Sadaoui Z, Boukhelata N, Azouaou N, Kaci Y (2009) Removal of cadmium from aqueous solution by polysaccharide produced from Paenibacillus polymyxa. J Hazard Mater 172(2–3):1150–1155. doi:10.1016/j.jhazmat.2009.07.116
Morillo J, Aguilera M, Ramos-Cormenzana A, Monteoliva-Sánchez M (2006) Production of a metal-binding exopolysaccharide by Paenibacillus jamilae using two-phase olive-mill waste as fermentation substrate. Curr Microbiol 53(3):189–193. doi:10.1007/s00284-005-0438-7
Morillo J, García-Ribera R, Quesada T, Aguilera M, Ramos-Cormenzana A, Monteoliva-Sánchez M (2008) Biosorption of heavy metals by the exopolysaccharide produced by Paenibacillus jamilae. World J Microbiol Biotechnol 24(11):2699–2704
Nwodo U, Green E, Okoh A (2012) Bacterial exopolysaccharides: functionality and prospects. Int J Mol Sci 13(11):14002–14015
Pattathil S, Harper A, Bar-Peled M (2005) Biosynthesis of UDP-xylose: characterization of membrane-bound AtUxs2. Planta 221(4):538–548. doi:10.1007/s00425-004-1471-7
Pollock T, van Workum W, Thorne L, Mikolajczak M, Yamazaki M, Kijne J, Armentrout R (1998) Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J Bacteriol 180(3):586–593
Raza W, Makeen K, Wang Y, Xu Y, Qirong S (2011) Optimization, purification, characterization and antioxidant activity of an extracellular polysaccharide produced by Paenibacillus polymyxa SQR-21. Bioresour Technol 102(10):6095–6103
Reckseidler-Zenteno SL (2012) Capsular polysaccharides produced by the bacterial pathogen Burkholderia pseudomallei. In: Karunaratne DN (ed) The complex world of polysaccharides. InTech, Rijeka, pp 127–152
Rodgers MW, Bolwell GP (1992) Partial purification of Golgi-bound arabinosyltransferase and two isoforms of xylosyltransferase from French bean (Phaseolus vulgaris L.). Biochem J 288(3):817–822
Ruan M, Yang Z, Zeng G, Xiong L, Tao R, Wang R, Liu Y (2007) Flocculating capability and mechanism of bioflocculant produced by Paenibacillus polymyxa GA1. Huanjing Kexue 28(10):2336–2341
Ryll T, Wagner R (1991) Improved ion-pair high-performance liquid chromatographic method for the quantification of a wide variety of nucleotides and sugar—nucleotides in animal cells. J Chromatogr B 570(1):77–88. doi:10.1016/0378-4347(91)80202-N
Sánchez-Andújar B, Coronado C, Philip-Hollingsworth S, Dazzo FB, Palomares AJ (1997) Structure and role in symbiosis of the exoB gene of Rhizobium leguminosarum bv trifolii. Mol Gen Genet 255(2):131–140. doi:10.1007/s004380050481
Sasaki Y, Ito Y, Sasaki T (2004) thyA as a selection marker in construction of food-grade host-vector and integration systems for streptococcus thermophilus. Appl Environ Microbiol 70(3):1858–1864. doi:10.1128/aem. 70.3.1858-1864.2004
Shimizu T, Matsusaka E, Nagakura N, Takayanagi K, Masuzawa T, Iwamoto Y, Morita T, Mifuchi I, Yanagihara Y (1987) Chemical properties of lipopolysaccharide-like substance (LLS) extracted from Leptospira interrogans serovar canicola strain Moulton. Microbiol Immunol 31(8):717
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1(9):784–791. doi:10.1038/nbt1183-784
Suzuki K, Watanabe K, Masumura T, Kitamura S (2004) Characterization of soluble and putative membrane-bound UDP-glucuronic acid decarboxylase (OsUXS) isoforms in rice. Arch Biochem Biophys 431(2):169–177. doi:10.1016/j.abb.2004.08.029
Tang J, Qi S, Li Z, An Q, Xie M, Yang B, Wang Y (2014) Production, purification and application of polysaccharide-based bioflocculant by Paenibacillus mucilaginosus. Carbohydr Polym 113(0):463–470. doi:10.1016/j.carbpol.2014.07.045
Wang C-L, Huang T-H, Liang T-W, Fang C-Y, Wang S-L (2011) Production and characterization of exopolysaccharides and antioxidant from Paenibacillus sp. TKU023. Biotechnology 28:559–565. doi:10.1016/j.nbt.2011.03.003
Wang Y, Li X, Milne CB, Janssen H, Lin W, Phan G, Hu H, Jin Y-S, Price ND, Blaschek HP (2013) Development of a gene knockout system using mobile group II introns (targetron) and genetic disruption of acid production pathways in Clostridium beijerinckii. Appl Environ Microbiol 79(19):5853–5863. doi:10.1128/aem. 00971-13
Wen Y, Wu X, Teng Y, Qian C, Zhan Z, Zhao Y, Li O (2011) Identification and analysis of the gene cluster involved in biosynthesis of paenibactin, a catecholate siderophore produced by Paenibacillus elgii B69. Environ Microbiol 13(10):2726–2737. doi:10.1111/j.1462-2920.2011.02542.x
Wu XC, Chen YM, Li YD, Li O, Zhu L, Qian CD, Tao XL, Teng Y (2011) Constitutive expression of Vitreoscilla haemoglobin in Sphingomonas elodea to improve gellan gum production. J Appl Microbiol 110(2):422–430. doi:10.1111/j.1365-2672.2010.04896.x
Yan J-K, Li L, Wang Z-M, Wu J-Y (2010) Structural elucidation of an exopolysaccharide from mycelial fermentation of a Tolypocladium sp. fungus isolated from wild Cordyceps sinensis. Carbohydr Polym 79(1):125–130. doi:10.1016/j.carbpol.2009.07.047
Yin G, Sun Z, Liu N, Zhang L, Song Y, Zhu C, Wen F (2009) Production of double-stranded RNA for interference with TMV infection utilizing a bacterial prokaryotic expression system. Appl Microbiol Biotechnol 84(2):323–333. doi:10.1007/s00253-009-1967-y
Zhu L, Wu X, Li O, Chen Y, Qian C, Teng Y, Tao X, Gao H (2011) Cloning and knockout of phytoene desaturase gene in Sphingomonas elodea ATCC 31461 for economic recovery of gellan gum. J Ind Microbiol Biotechnol 38(9):1507–1513. doi:10.1007/s10295-010-0937-9
Zhu L, Wu X, Li O, Qian C, Gao H (2012) Cloning and characterization of genes involved in nostoxanthin biosynthesis of Sphingomonas elodea ATCC 31461. PLoS ONE 7(4):e35099. doi:10.1371/journal.pone.0035099
Acknowledgments
The study was supported by the Major State Basic Research Development Program (973 Program, Grant no. 2010CB833803).
Conflict of interest
The authors declare that there are no conflicts of interest. We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ou Li and Chao-Dong Qian contributed equally to the development of this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 670 kb)
Rights and permissions
About this article
Cite this article
Li, O., Qian, CD., Zheng, Dq. et al. Two UDP-glucuronic acid decarboxylases involved in the biosynthesis of a bacterial exopolysaccharide in Paenibacillus elgii . Appl Microbiol Biotechnol 99, 3127–3139 (2015). https://doi.org/10.1007/s00253-014-6362-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6362-7