Abstract
In response to danger signals, macrophages rapidly produce many inflammatory cytokines that trigger the cascade release of inflammatory mediators, leading to tissue damage, which is an important cause of clinical manifestations of syphilis at all stages. However, we still know very little about the specific mechanism of this process. Tp0768 is an infection-stage–dependent antigen that plays an important role in the infection of Treponema pallidum. In this study, we demonstrated that Tp0768 stimulation of macrophages can cause IL-1β, IL-6, and IL-8 mRNA expression levels to increase in a dose- and time-dependent manner. Further research showed that Tp0768 activated ER stress and the ROS/NF-κB pathway in macrophages. Inhibition of ER stress and the ROS/NF-κB pathway inhibited the expression of IL-1β, IL-6, and IL-8 induced by Tp0768. In addition, pretreatment with a PERK pathway inhibitor significantly reduced the expression of the NF-κB and JNK pathways, while also downregulating the expression of IL-1β, IL-6, and IL-8. Tp0768 stimulation can activate IRE1α/XBP-1 signaling and participate in the induction of inflammatory cytokines through the JNK pathway. These findings indicate that Tp0768 promotes the secretion of proinflammatory cytokines IL-1β, IL-6, and IL-8 by macrophages through ER stress and the ROS/NF-κB pathway, which are also involved in the activation of the NF-κB and JNK pathways that are induced by the PERK pathway and activation of IRE1α/XBP-1 signaling.
Key points
• This study found for the first time that the recombinant Treponema pallidum protein Tp0768 promotes the production of IL-1β, IL-6, and IL-8 by macrophages through ER stress.
• Recombinant Treponema pallidum protein Tp0768 regulates the ROS/NF-κB pathway through ER stress.
• ER stress-related pathway PERK induces the expression of IL-1β, IL-6, and IL-8 by activating the NF-κB pathway and the JNK pathway.
• IRE1α can induce the splicing of XBP-1mRNA and activate the JNK pathway.
Similar content being viewed by others
Introduction
Syphilis is a chronic sexually transmitted disease caused by Treponema pallidum (T. pallidum) infection (Luo et al. 2018). Its clinical manifestations are complex, and infection can invade various organs throughout the body, causing damage to multiple organs. The inflammatory response caused by T. pallidum replication in tissues and the body’s immune response towards pathogens are the causes of the clinical manifestations at all stages of syphilis (Peeling et al. 2017; Li et al. 2020a). Although simple kits are available for diagnostics and penicillin is an effective drug used for treatment, syphilis has begun to be a resurrected global public health problem in recent years (Hook 2017).
The endoplasmic reticulum (ER) is a specialized perinuclear organelle responsible for secretion, biosynthesis of transmembrane proteins, as well as the modification and folding of proteins to maintain protein homeostasis (Moon et al. 2018; Li et al. 2020b). The change in ER homeostasis leads to the accumulation of unfolded proteins in the ER cavity, which leads to ER stress activation (Hu et al. 2018). As one of the major protein quality control systems, ER stress consists of a complex network of three signal transmission pathways, each of which involves a protein sensor: activated transcription factor 6 (ATF6), inositol requiring enzyme 1 (IRE1), or protein kinase RNA (PKR)–like kinase (PERK) (Rashid et al. 2015). Under basic/unstressed conditions, the protein partner, BiP/GRP78, located in ER binds to the cavity domains of these protein sensors to maintain the inactive state of these pathways. When ER is stressed, the accumulation of incorrectly folded proteins and increased protein load in ER causes BiP/GRP78 to dissociate from these UPR sensors and bind to unfolded or misfolded proteins, which activates PERK, IRE1 and ATF6 (Grover et al. 2018). In chronic diseases, ER stress may contribute to the production of persistent inflammatory mediators, thus hindering the in vivo balance mechanism. Increasing evidence shows that ER stress is a powerful controller of cell response to external stimuli and inflammatory signal transmission and is an important mechanism that determines cell fate (Muriach et al. 2014; Chaudhari et al. 2014). ER stress can be activated by a variety of pathogens, including viruses such as Candida albicans, Chlamydia pneumoniae, and Mycobacterium tuberculosis (Li et al. 2018; Walenna et al. 2020; Liang et al. 2019). Early ER stress is a defense response triggered by specific stimulatory conditions, which aims to protect cell vitality and normal physiological function, but it is destructive beyond physiological control and seriously affects immune response and inflammation.
Tp0768 is considered to be a T. pallidum infection-dependent antigen (Gerber et al. 1996). Previous studies have shown that high immunogenic Tp0768 (also known as TpN44.5 or TmpA) showed high sensitivity and specificity for the serological diagnosis of syphilis (Liu et al. 2019; Backhouse and Nesteroff 2001). However, the pathogenesis of Tp0768 is still unclear. In this study, we studied the inflammatory effect of Tp0768 on macrophages and the important role of ER stress and the ROS/NF-κB pathway in this process in vitro. Understanding Tp0768-induced immune regulation will help elucidate the pathogenesis of innate immunity and subsequent adaptation mechanisms, which may be related to the treatment of syphilis and the development of vaccines.
Materials and Method
Chemicals and reagents
DMSO, phorbol 12-myristate 13-acetate (PMA), N-acetyl-l-cysteine (NAC), and 4-phenylbutric acid (4-PBA) were purchased from Sigma-Aldrich. STF-083010, GSK2656157, and SP600125 were purchased from MedChemExpress. Pyrrolidine dithiocarbamate (PDTC) was purchased from Beyotime. Antibodies against Bip, PERK, IRE1α, NF-κB p65, JNK, P-JNK, and GAPDH and secondary antibodies were purchased from Cell Signaling Technology.
Expression and purification of Tp0768
Recombinant plasmids were constructed using T. pallidum Nichols strain genomic DNA as a template and were transformed into Escherichia coli for expression. After purification of the protein, endotoxin was removed and detected using an endotoxin removal kit and a limulus reagent detection kit, respectively. The endotoxin content in the Tp0768 formulation used in the experiments was less than 0.1 EU/mL.
Cell culture
THP-1 cells and Raw264.7 cells were purchased from the American Type Culture Collection and cultured in RPMI 1640 and Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, respectively. Cell growth conditions were maintained at 37 °C and 5% CO2; 100 ng/mL PMA was used to induce THP-1 cells to differentiate into macrophages for 24 h to be used in the experiments.
RT-qPCR
Total RNA was extracted using TRNzol-A+ Reagent. cDNA was synthesized using the Fastking RT Kit, by following the manufacturer’s instructions. All primer designs were synthesized by Sangon Biotech. The primer sequences are listed in Table 1. Real-time PCR was performed in the Light-Cycler 480 System using Super Real Premix Plus (SYBR Green). GAPDH was used as an internal standardized control. The relative gene expression level was quantified by normalizing it to the endogenous GAPDH expression level using the 2−∆∆CT method.
Transmission electron microscopy
The cells were pre-fixed using an electron microscope fixing solution for 2 h and washed 3 times with PBST. Fixation was conducted using 1% osmium tetroxide for 2 h, followed by dehydration using ethanol. After being embedded overnight, it was cut into ultra-thin sections and observed under a transmission electron microscope at an acceleration voltage of 100 kV.
Western blotting analysis
After harvesting the cells, each well was lysed using a RIPA buffer containing a protease and phosphatase inhibitor. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate an equal amount of protein from the cells and was transferred onto PVDF membranes. The membrane was blocked using a 5% skimmed milk powder buffer for 1 h at room temperature. After blocking, the membrane was incubated with suitable primary antibodies: Bip (1:1000), PERK (1:1000), IRE1α (1:1000), NF-κB p65 (1:1000), JNK (1:1000), P-JNK (1:1000), and GAPDH (1:5000) at 4 °C overnight, and then incubated with a secondary antibody (1:2000) for 1.5 h at room temperature. The resulting bands were visualized using BeyoECL Star. ImageJ software was used to quantify band intensity and normalize band intensity using GAPDH.
Detection of ROS
Following instructions given by the active oxygen detection kit manufacturer, cells were collected, the fluorescent probe, DCFH-DA, was loaded and incubated in a 37 °C cell incubator for 20 min. After washing three times with serum-free medium, flow cytometry was used to detect fluorescence. The detection parameters were set to an excitation wavelength of 488 nm and emission wavelength of 525 nm. Raw264.7 cells were captured using a fluorescence microscope, and the average fluorescence intensity was quantified using ImageJ software.
Cellular immunofluorescence
Cells were seeded onto polylysine-coated slides. The collected cells were fixed using 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 and subsequently closed. A diluted NF-κB p65 antibody (1:300) was added into each well and incubated at 4 °C overnight. After washing with PBST, the cells were incubated with goat anti-rabbit IgG H&L (Alexa Fluor® 488) for 1 h. Nuclei were counterstained with DAPI. The slides were mounted using an anti-fluorescence quenching mount. A laser confocal scanning microscope was used to capture fluorescence images.
Determination of Xbp-1 splicing
Cells were seeded into 12-well plates and were pretreated with different concentrations of STF-083010 for 1 h, and then co-cultured with Tp0768 for 24 h. The cells were collected, and total RNA was extracted. After reverse transcription into cDNA, Super Real Premix Plus (SYBR Green) was used to perform real-time PCR analysis in a Light-Cycler 480 System. The primer sequences are listed in Table 1. The PCR products were electrophoresed on a 2% agarose gel for 20 min at a voltage of 50 V.
siRNA transfection
XBP-1s and negative control siRNA were purchased from HonorGene (Changsha, China). According to the manufacturer’s protocol, Lipofectamine 2000 (ThermoFisher Scientific) was used to transfect siRNA into cells.
Statistical analysis
Statistical analyses were performed using SPSS software or GraphPad Prism 7. One-way ANOVA was used to analyze more than 3 groups of data, and Student’s t test was used to analyze two independent groups. The results are presented as mean ± SD of experiments performed in triplicate and p value of < 0.05 was considered to be statistically significant.
Results
Tp0768 induced the production of inflammatory cytokines in macrophages
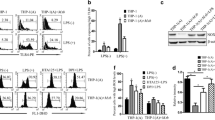
In order to analyze the effect of Tp0768 on the expression of IL-1β, IL-6, and IL-8 in macrophages, we incubated the macrophages with different concentrations of Tp0768 for 24 h. Tp0768 significantly increased IL-1β, IL-6, and IL-8 mRNA expression in a dose-dependent manner (Fig. 1a, c). Based on the above results, 5 μg/mL Tp0768 was used for macrophage incubation for 0, 6, 12, 24, 48, and 72 h. IL-1β, IL-6, and IL-8 mRNA levels increased significantly at 24 h, in a time-dependent manner (Fig. 1b, d).
Tp0768 induced the expression of IL-1β, IL-6, and IL-8 in THP-1–differentiated macrophages and Raw264.7 cells. The mRNA expression of IL-1β, IL-6, and IL-8 in THP-1–differentiated macrophages (a) and Raw264.7 cells (c) was measured using RT-qPCR after treatment with Tp0768 at different concentrations for 24 h. b, d Five micrograms per milliliter Tp0768-treated cells at different time (0, 6, 12, 24, 48, and 72 h) were used to determine the mRNA expression of IL-1β, IL-6, and IL-8 using RT-qPCR. Values are expressed as fold changes relative to GAPDH-normalized mRNA levels. All data are presented as mean ± SD of at least three independent experiments. NS, not significant. *p < 0.05, **p < 0.01, and ***p < 0.001 indicate a significant difference from the control group
Inhibition of ER stress reduced inflammatory cytokine production induced by Tp0768
In order to confirm whether Tp0768 induced ER stress, we used transmission electron microscopy to observe endoplasmic reticulum expansion; swelling and the normal folding structure disappeared after Tp0768 stimulation, compared with the control group (Fig. 2a). In order to clarify this further, we examined representative molecular markers of ER stress. After stimulation of cells with Tp0768, the expression levels of PERK, Bip, and IRE1α increased significantly and were dose dependent (Fig. 2b; Fig. S1a, b). These results indicate that Tp0768 can induce ER stress in THP-1–differentiated macrophages and Raw264.7 cells. In order to verify the relationship between ER stress and inflammation, we used the ER stress inhibitor 4-PBA to block the UPR pathway. As shown (Fig. 2c; Fig. S1), 2.5 mM 4-PBA significantly reduced the expression of PERK, Bip, and IRE1α. In addition, 4-PBA intervention was able to effectively decrease Tp0768-induced IL-1β, IL-6, and IL-8 mRNA levels (Fig. 2d, e).
Tp0768 mediated the expression of IL-1β, IL-6, and IL-8 through ER stress. a Electron microscope image of the ultrastructure of THP-1–differentiated macrophages and Raw264.7 cells treated with 5 μg/mL Tp0768. The high-magnification image highlights the swollen endoplasmic reticulum. b After treatment of cells with different concentrations of Tp0768 for 24 h, ER stress-related proteins, PERK, IRE1α, and Bip, were detected using western blotting. c Cells pre-incubated with 4-PBA (5, 2.5, and 1.25 mM) for 1 h were treated with Tp0768 for 24 h, and the protein levels of PERK, Bip, and IRE1 were evaluated using western blotting. d, e After pretreatment with 4-PBA (2.5 mM) and co-treatment with Tp0768 for 24 h, the mRNA expression of IL-1β, IL-6, and IL-8 were evaluated using RT-qPCR. Values are expressed as fold changes relative to GAPDH-normalized mRNA levels. All data are presented as mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 indicate a significant difference from the control group
The induction of cytokines by Tp0768 is regulated by the ROS/NF-κB pathway
Cell oxidative stress is an endogenous regulator that determines cell fate. Studies have shown that ROS accumulation can activate redox signaling pathways, some of which often cross-talk with ER stress and inflammation (Dehdashtian et al. 2018; Chen et al. 2019). We hypothesized that the accumulation of ROS induced by Tp0768 could be suppressed by ER stress inhibition. In order to validate our assumption, we used flow cytometry and cellular immunofluorescence to detect ROS levels in THP-1 differentiated macrophages and Raw264.7 cells, respectively. The results showed that Tp0768 stimulation could significantly induce intracellular ROS production, and that 4-PBA and NAC intervention could significantly decrease intracellular ROS levels induced by Tp0768 (Fig. 3a–d). In addition, we detected NF-κBp65 signaling using immunofluorescence staining and western blotting. As shown (Fig. 4a, b), when macrophages were exposed to Tp0768, the protein level of NF-κBp65 was upregulated in a dose-dependent manner and nuclear translocation occurred. Treatment of macrophages with Tp0768 in the presence of the antioxidant, NAC, showed that NAC treatment inhibited the increase in NF-κBp65 protein levels in macrophages induced by Tp0768 (Fig. 4c). In order to further confirm whether NF-κB is closely related to Tp0768-induced IL-1β, IL-6, and IL-8 expression, we co-treated cells with the NF-κB inhibitor, PDTC, and Tp0768 for 24 h. The secretion of IL-1β, IL-6, and IL-8 in Tp0768-treated macrophages significantly increased, compared to the control group, while the secretion decreased after treatment with PDTC, indicating that inhibition of the NF-κB pathway decreased IL-1β, IL-6, and IL-8 expression induced by Tp0768 (Fig. 4d, e). Taken together, these results indicate that Tp0768 regulates IL-1β, IL-6, and IL-8 production through the ROS/NF-κB pathway in ER-stressed cells.
Tp0768-induced ROS release. a THP-1–differentiated macrophages were treated with Tp0768 in the absence or presence of NAC (5 mM) or 4-PBA (2.5 mM) for 1 h, and the ROS level in the cells were determined using flow cytometry. b Quantitative analysis of the average fluorescence intensity of ROS using FlowJo software. c Analysis of ROS production in Raw264.7 cells using cellular immunofluorescence (× 200). d Quantification of ROS levels using ImageJ software. *p < 0.05, **p < 0.01, and ***p < 0.001 indicate a significant difference
Tp0768 regulated the production of proinflammatory cytokines in cells through the ROS/NF-κB pathway. a NF-κB (p65) was detected using western blotting, and its gray value was analyzed using ImageJ software. b Nuclear translocation of NF-κB (p65) was assessed using immunofluorescence staining and confocal microscopy. DIPA was used for nuclear staining at a scale of 5 μm. c NF-κB (p65) levels were detected using western blotting in the absence or presence of NAC. The relative mRNA levels of IL-1β, IL-6, and IL-8 in THP-1–differentiated macrophages (d) and Raw264.7 cells (e) were detected using RT-PCR in the absence or presence of PDTC, respectively. Values are expressed as fold changes relative to GAPDH normalized mRNA levels. All data are presented as mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 indicate a significant difference from the control group
PERK induced inflammatory cytokine gene expression through the JNK and NF-κB pathways
Current research indicates that the ER stress-induced inflammatory response proceeds mainly via the PERK and IRE1α pathways. In order to determine whether the UPR-related pathway, PERK, is involved in THP-1–differentiated macrophage and Tp0768-induced IL-1β, IL-6, and IL-8 expression in Raw264.7 cells, we used a PERK inhibitor, GSK2656157, to pretreat the cells. After the cells were treated with different concentrations of GSK2656157 and Tp0768 for 24 h, the results showed that when the PERK pathway was inhibited, the expression of NF-κB (P65) and P-JNK decreased significantly, and IL-1β, IL-6, and IL-8 mRNA expression were also downregulated (Fig. 5a–d). In order to further clarify the effect of the JNK pathway on the expression of inflammatory genes, we determined the expression of inflammatory genes 24 h after treatment with Tp0768 in the presence of different concentrations of SP600125 (a selective inhibitor of the JNK pathway). The results showed that SP600125 attenuated the expression of IL-1β, IL-6, and IL-8 mRNA induced by Tp0768 (Fig. 5c, d). In summary, PERK-activated JNK and NF-κB pathways are involved in the induction of inflammatory genes.
PERK induced the expression of inflammatory cytokine genes through the JNK and NF-κB pathways. a, b Cells were pretreated with GSK2656157 (5, 10, and 20 μm) for 1 h, then treated with 5 μg/mL Tp0768 for 24 h, and PERK, P65, JNK, and P-JNK were detected using western blotting. c, d Relative mRNA levels of IL-1β, IL-6, and IL-8 using RT-PCR in the absence or presence of GSK2656157 or SP600125 (10 μm). Values are expressed as fold changes relative to GAPDH-normalized mRNA levels. Data in histograms are presented as mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 indicate a significant difference from the control group
IRE1α/XBP-1 induced inflammatory cytokine gene expression through the JNK pathway
IRE1 is an important sensor of ER stress, and its activation of XBP-1 splicing plays an important role in ER stress-induced inflammation. In order to determine whether the IRE1-XBP-1 pathway is activated in THP-1–differentiated macrophages and Raw264.7 cells in response to Tp0768 infection, we first evaluated the splicing effect of IRE1α on XBP-1. After co-culture of the cells with different concentrations of STF-083010, the cells were treated with Tp0768 for 24 h, and the RT-qPCR products were separated using agarose gel electrophoresis. As shown (Fig. 6a, b), compared with the control group, Tp0768 infection significantly promoted the splicing of XBP-1 mRNA and the production of XBP-1s; 20 μM STF-083010 intervention was able to significantly inhibit the splicing of XBP-1 mRNA induced by Tp0768. In addition, we investigated whether JNK signaling is activated through IRE1α/XBP-1 signaling and is involved in Tp0768-induced inflammation. The results indicated that the inhibition of IRE1α signaling decreased phosphorylated JNK levels (Fig. 6c, d). Silencing the expression of XBP-1s with siRNA inhibited the expression of phosphorylated JNK in cells, which indicated that JNK is activated via the IRE1α/XBP-1 pathway (Fig. 6e, f). When SP600125 was used to inhibit JNK kinase activity, the transcription of IL-1β, IL-6, and IL-8 were significantly suppressed (Fig. 6g, h). These results indicate that IRE1α/XBP-1 is involved in Tp0768-induced inflammatory gene expression through the JNK pathway.
EIF/XBP1 induced the expression of inflammatory cytokine genes through the JNK pathway. a, b Cells were pretreated with STF-083010 (10, 20, and 40 μm) for 1 h, and then treated with 5 μg/mL Tp0768 for 24 h. The XBP1 gene was amplified using PCR and analyzed through separation on a 2% agarose gel. Xbp-1s, spliced form; Xbp-1u, unspliced form. c, d The expression levels of IRE1α, JNK, and P-JNK were detected using western blotting. e, f After transfection with control siRNA or XBP-1s siRNA for 48 h, the expression level of XBP-1s mRNA was detected by qRT-PCR and the expression level of P-JNK protein was detected by Western blotting. The relative mRNA levels of IL-1β, IL-6, and IL-8 in the THP-1–differentiated macrophages (g) and Raw264.7 cells (h) were detected using RT-PCR. Values are expressed as fold changes relative to GAPDH-normalized mRNA levels. The data in the histograms are presented as mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 indicate a significant difference from the control group
Discussion
Compared with the large amount of information on the pathogenic mechanism of other bacterial pathogens, our understanding of the pathogenic mechanism of syphilis lags far behind that of other common bacterial infections because T. pallidum is difficult to be cultured in vitro for a long period and genetic techniques cannot be used to describe the determinants of its virulence. Although the whole genome of T. pallidum has been sequenced, only research aimed at annotating the functions of T. pallidum pathogenic proteins has been carried out so far (Staudova et al. 2014; Petrosova et al. 2012; Weinstock et al. 1998). At present, research on the function of T. pallidum pathogenic proteins is mainly on outer membrane proteins, and our understanding of lipoprotein functions is still limited. Lipoproteins constitute the highest content of proteins in T. pallidum and usually play a decisive role in the mechanism of T. pallidum and its environment. Helicoid lipoprotein is a proinflammatory molecule that can regulate innate and adaptive immune responses (Kelesidis 2014). Understanding lipoprotein-induced immune regulation will help elucidate the pathogenesis of innate immunity and subsequent adaptation mechanisms, which may be related to inflammation and related vaccine development in spirochetes disease. Studies have shown that lipoprotein Tp0768 is an infection-dependent antigen that has a high value for the clinical diagnosis of syphilis serology (Liu et al. 2019; Runina et al. 2018). However, little is known about the role of Tp0768 in inflammation.
Previous studies have shown that the innate immune response of phagocytes towards microorganisms depend on the core signaling cascade downstream of the pattern recognition receptor (PRR), which leads to an adaptive immune response that produces host resistance towards infection and induces the production of inflammatory cytokines (Moretti and Blander 2017). Autonomic cellular responses aimed at ensuring cell homeostasis, such as autophagy, metabolic stress, ER stress, and mitochondrial stress, are important for the mechanism of innate immunity. ER stress/UPR is a conservative adaptive mechanism developed by eukaryotic cells to remove unfolded proteins and restore ER homeostasis. When ER stress cannot be reversed, cell dysfunction eventually leads to cell death. However, ER stress caused by microbial infection is usually long term. ER stress has been proven to be associated with inflammatory diseases caused by infections of various microorganisms, such as acute viral myocarditis, chronic hepatitis C virus, tuberculosis, and placental inflammation (Cai et al. 2015; Dash et al. 2019; Lim et al. 2015; Byndloss et al. 2019). In order to verify whether Tp0768 induces macrophages to produce ER stress, we observed the ultrastructure of ER using transmission electron microscopy (TEM) and found many swollen and irregular ER. Western blotting used to confirm the UPR-related proteins also showed that Tp0768 induced ER stress in macrophages in a dose-dependent manner. Therefore, our results show that Tp0768 induced macrophages to produce ER stress that may play a crucial role in the pathogenesis of T. pallidum.
Genes regulated by the NF-κB transcription factor family are involved in a variety of physiological and pathological processes, including inflammation, immune response, autophagy, and apoptosis (Tam et al. 2012). In this study, we confirmed that Tp0768 can promote the activation of NF-κB through the PERK pathway, thereby inducing the expression of proinflammatory cytokines IL-1β, IL-6, and IL-8 in macrophages. Previous studies have also shown that the NF-κB pathway plays an important role in T. pallidum–mediated inflammation. For example, T. pallidum induces IL-6, MCP-1, and ICAM-1 in human dermal vascular smooth muscle cells through the NF-κB signaling pathway expression, thereby promoting the migration and adhesion of THP-1 cells to human dermal vascular smooth muscle cells (Gao et al. 2019). Tp92 recognizes CD14 and TLR2 and transfers signals to the downstream pathway NF-κB to mediate IL-8 production (Luo et al. 2018). In response to ER stress, PERK-eIF2α-mediated weakening of translation can directly promote NF-κB activation (Kitamura 2011). Because the half-life of IκB is much shorter than the half-life of NF-κB, attenuated translation increases the ratio of NF-κB to IκB, which causes NF-κB to translocate to the nucleus in response to ER stress (Zhang and Kaufman 2008). However, the specific mechanism of Tp0768 inducing NF-κB activation through PERK pathway in macrophages needs further study.
IRE1α/XBP-1 is the most conservative branch of evolution in ER stress (Brenner et al. 2013). After dissociation from Bip, activated IRE1α activates RNase activity after undergoing homodimerization and autophosphorylation. XBP-1 mRNA was cleaved to XBP-1s mRNA by an unconventional splicing mechanism that removed 26 nucleotide introns and translated into functional active transcription factors to induce the expression of UPR target inflammatory genes (Li et al. 2019a, b). Our study found that Tp0768 induces XBP-1 splicing through IRE1α. In addition, studies have shown that the cytoplasmic portion of IRE1 can bind to TRAF2, which acts as an adapter protein to couple plasma membrane receptors to JNK activation (Urano et al. 2000). And a recent report shows that T. pallidum flagellin stimulates MMP-13 expression through the JNK signaling pathway in human epidermal keratinocytes, which is conducive to the induction of skin inflammation and pathogen invasion (Jiang et al. 2017). Our data indicate that activation of IRE1α can recruit protein kinase JNK, thereby activating JNK and inducing the expression of IL-1β, IL-6, and IL-8. These findings provide strong evidence that Tp0768 activates the IRE1α-XBP-1-JNK signaling axis leading to secretion of inflammatory factors.
As a part of basic metabolic function, ROS is mainly produced by various enzymatic reactions and specific enzymes, such as NOX, in cell compartments, and plays a signaling function in cells for the regulation of cell proliferation and survival (D'Autreaux and Toledano 2007; El-Kenawi and Ruffell 2017). ROS levels above a certain level can damage cell components, interfere with normal cell processes, and lead to cell death, thus leading to metabolic dysfunction and inflammatory response (Burgos-moron et al. 2019). One of the most significant characteristics of Tp0768-treated macrophages is the production of ROS. Under pathological conditions, ER is an important compartment that produces ROS (Cao and Kaufman 2014). ERO1 transfers electrons from PDI to molecular oxygen. Ero1p bound by FAD oxidizes PDI and then directly oxidizes folding proteins (Mennerich et al. 2019; Zeeshan et al. 2016). Ero1p bound by FAD transfers electrons to molecular oxygen, resulting in ROS generation (Tu and Weissman 2004). In order to study whether ROS play a potential role in inflammatory response, we used 4-PBA to inhibit ER stress and found that the production of ROS decreased, indicating that Tp0768 may induce the accumulation of ROS through ER stress. In addition, we also examined the activation of NF-κB, an important downstream pathway, in which ROS act as an inflammatory signal modulator. Our results show that Tp0768 induced activation and translocation of NF-κ B. Antioxidant NAC was able to inhibit the expression of NF-κBp65 in macrophages induced by Tp0768. In conclusion, these results indicated that Tp0768-induced ER stress plays a significant role in inflammatory response through the ROS/NF-κB axis.
In view of the central role of ER stress in the management of cell stress response in endoplasmic reticulum homeostasis, many pathogens have been reported to activate or inhibit ER stress to promote their own survival. For example, chlamydia can inhibit ER stress during infection, reduce the phosphorylation level of eIF2α, block the translocation of CHOP protein to the nucleus, and increase the bacterial load (Mehlitz et al. 2014). In addition, similar to our experimental results, the secretory effector protein VceC of Brucella abortus activates IRE1, and its activation can occur simultaneously with the expression of IRE1-dependent proinflammatory cytokines (De Jong et al. 2013). Different from our results, VceC induces the expression of proinflammatory cytokines through IRE1 activation of NOD1/NOD2 innate immune signals (Keestra-Gounder et al. 2016).
In general, this study confirmed for the first time that T. pallidum induces the secretion of proinflammatory cytokines by activating cellular ERS and its downstream pathways through its own effector protein (Fig. 7). We found that the PERK and IRE1α pathways of ER stress are significantly activated and promote the production of inflammatory cytokines through the activation of the NF-κB and JNK pathways. In addition, ER stress induced by Tp0768 can significantly increase ROS generation. Understanding the importance of ER stress and the related mechanisms of its proinflammatory effect may help to provide a new potential therapeutic strategy to control systemic inflammatory response of syphilis patients.
References
Backhouse JL, Nesteroff SI (2001) Treponema pallidum western blot: comparison with the FTA-ABS test as a confirmatory test for syphilis. Diagn Microbiol Infect Dis 39(1):9–14. https://doi.org/10.1016/s0732-8893(00)00213-3
Brenner C, Galluzzi L, Kepp O, Kroemer G (2013) Decoding cell death signals in liver inflammation. J Hepatol 59(3):583–594. https://doi.org/10.1016/j.jhep.2013.03.033
Burgos-Moron E, Abad-Jimenez Z, Maranon AM, Iannantuoni F, Escribano-Lopez I, Lopez-Domenech S, Salom C, Jover A, Mora V, Roldan I, Sola E, Rocha M, Victor VM (2019) Relationship between oxidative stress, ER stress, and inflammation in type 2 diabetes: the battle continues. J Clin Med 8(9). https://doi.org/10.3390/jcm8091385
Byndloss MX, Tsai AY, Walker GT, Miller CN, Young BM, English BC, Seyffert N, Kerrinnes T, de Jong MF, Atluri VL, Winter MG, Celli J, Tsolis RM (2019) Brucella abortus Infection of placental trophoblasts triggers endoplasmic reticulum stress-mediated cell death and fetal loss via type IV secretion system-dependent activation of CHOP. MBio 10(4). https://doi.org/10.1128/mBio.01538-19
Cai Z, Shen L, Ma H, Yang J, Yang D, Chen H, Wei J, Lu Q, Wang DW, Xiang M, Wang J (2015) Involvement of endoplasmic reticulum stress-mediated C/EBP homologous protein activation in coxsackievirus B3-induced acute viral myocarditis. Circ Heart Fail 8(4):809–818. https://doi.org/10.1161/circheartfailure.114.001244
Cao SS, Kaufman RJ (2014) Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal 21(3):396–413. https://doi.org/10.1089/ars.2014.5851
Chaudhari N, Talwar P, Parimisetty A, Lefebvre d’Hellencourt C, Ravanan P (2014) A molecular web: endoplasmic reticulum stress, inflammation, and oxidative stress. Front Cell Neurosci 8:213. https://doi.org/10.3389/fncel.2014.00213
Chen X, Guo X, Ge Q, Zhao Y, Mu H, Zhang J (2019) ER Stress activates the NLRP3 inflammasome: a novel mechanism of atherosclerosis. Oxidative Med Cell Longev 2019:3462530. https://doi.org/10.1155/2019/3462530
Dash S, Aydin Y, Wu T (2019) Integrated stress response in hepatitis C promotes Nrf2-related chaperone-mediated autophagy: a novel mechanism for host-microbe survival and HCC development in liver cirrhosis. Semin Cell Dev Biol. https://doi.org/10.1016/j.semcdb.2019.07.015
D'Autreaux B, Toledano MB (2007) ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 8(10):813–824. https://doi.org/10.1038/nrm2256
De Jong MF, Starr T, Winter MG, den Hartigh AB, Child R, Knodler LA, van Dijl JM, Celli J, Tsolis RM (2013) Sensing of bacterial type IV secretion via the unfolded protein response. mBio 4(1):e00418–e00412. https://doi.org/10.1128/mBio.00418-12
Dehdashtian E, Mehrzadi S, Yousefi B, Hosseinzadeh A, Reiter RJ, Safa M, Ghaznavi H, Naseripour M (2018) Diabetic retinopathy pathogenesis and the ameliorating effects of melatonin; involvement of autophagy, inflammation and oxidative stress. Life Sci 193:20–33. https://doi.org/10.1016/j.lfs.2017.12.001
El-Kenawi A, Ruffell B (2017) Inflammation, ROS, and mutagenesis. Cancer Cell 32(6):727–729. https://doi.org/10.1016/j.ccell.2017.11.015
Gao ZX, Liu LL, Lin LR, Tong ML, Liu F, Yang TC (2019) Treponema pallidum Induces the secretion of HDVSMC inflammatory cytokines to promote the migration and adhesion of THP-1 Cells. Front Cell Infect Microbiol 9:220. https://doi.org/10.3389/fcimb.2019.00220
Gerber A, Krell S, Morenz J (1996) Recombinant Treponema pallidum antigens in syphilis serology. Immunobiology 196(5):535–549
Grover S, Sharma T, Singh Y, Kohli S, P M, Singh A, Semmler T, Wieler LH, Tedin K, Ehtesham NZ, Hasnain SE (2018) The PGRS domain of Mycobacterium tuberculosis PE_PGRS protein Rv0297 is involved in endoplasmic reticulum stress-mediated apoptosis through toll-like receptor 4. MBio 9(3). https://doi.org/10.1128/mBio.01017-18
Hook EW 3rd (2017) Syphilis. Lancet 389(10078):1550–1557. https://doi.org/10.1016/s0140-6736(16)32411-4
Hu H, Tian M, Ding C, Yu S (2018) The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front Immunol 9:3083. https://doi.org/10.3389/fimmu.2018.03083
Jiang C, Xu M, Kuang X, Xiao J, Tan M, Xie Y, Xiao Y, Zhao F, Wu Y (2017) Treponema pallidum flagellins stimulate MMP-9 and MMP-13 expression via TLR5 and MAPK/NF-kappaB signaling pathways in human epidermal keratinocytes. Exp Cell Res 361(1):46–55. https://doi.org/10.1016/j.yexcr.2017.09.040
Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chávez-Arroyo A, Tsai AY, Cevallos SA, Winter MG, Pham OH, Tiffany CR, de Jong MF, Kerrinnes T, Ravindran R, Luciw PA, McSorley SJ, Bäumler AJ, Tsolis RM (2016) NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532(7599):394–397. https://doi.org/10.1038/nature17631
Kelesidis T (2014) The cross-talk between spirochetal lipoproteins and immunity. Front Immunol 5(310). https://doi.org/10.3389/fimmu.2014.00310
Kitamura M (2011) Control of NF-κB and inflammation by the unfolded protein response. Int Rev Immunol 30(1):4–15. https://doi.org/10.3109/08830185.2010.522281
Li J, Yu Q, Zhang B, Xiao C, Ma T, Yi X, Liang C, Li M (2018) Stress-associated endoplasmic reticulum protein 1 (SERP1) and Atg8 synergistically regulate unfolded protein response (UPR) that is independent on autophagy in candida albicans. Int J Med Microbiol 308(3):378–386. https://doi.org/10.1016/j.ijmm.2018.03.004
Li Y, Jiang W, Niu Q, Sun Y, Meng C, Tan L, Song C, Qiu X, Liao Y, Ding C (2019a) eIF2α-CHOP-BCl-2/JNK and IRE1α-XBP1/JNK signaling promote apoptosis and inflammation and support the proliferation of Newcastle disease virus. Cell Death Dis 10(12):891. https://doi.org/10.1038/s41419-019-2128-6
Li R, Liu T, Shi J, Luan W, Wei X, Yu J, Mao H, Liu P (2019b) ROR2 induces cell apoptosis via activating IRE1α/JNK/CHOP pathway in high-grade serous ovarian carcinoma in vitro and in vivo. J Transl Med 17(1):428. https://doi.org/10.1186/s12967-019-02178-x
Li W, Luo C, Xie X, Xiao Y, Zhao F, Cai J, Zhou X, Zeng T, Fu B, Wu Y, Xiao X, Liu S (2020a) Identification of key genes and pathways in syphilis combined with diabetes: a bioinformatics study. AMB Express 10(1):83. https://doi.org/10.1186/s13568-020-01009-3
Li W, Cao T, Luo C, Cai J, Zhou X, Xiao X, Liu S (2020b) Crosstalk between ER stress, NLRP3 inflammasome, and inflammation. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-020-10614-y
Liang S, Wang F, Bao C, Han J, Guo Y, Liu F, Zhang Y (2019) BAG2 ameliorates endoplasmic reticulum stress-induced cell apoptosis in Mycobacterium tuberculosis-infected macrophages through selective autophagy. Autophagy:1–15. https://doi.org/10.1080/15548627.2019.1687214
Lim YJ, Choi JA, Lee JH, Choi CH, Kim HJ, Song CH (2015) Mycobacterium tuberculosis 38-kDa antigen induces endoplasmic reticulum stress-mediated apoptosis via toll-like receptor 2/4. Apoptosis 20(3):358–370. https://doi.org/10.1007/s10495-014-1080-2
Liu W, Deng M, Zhang X, Yin W, Zhao T, Zeng T, Liu S, Xiao Y, Zhang L, Luo X, Zhao F (2019) Performance of novel infection phase-dependent antigens in syphilis serodiagnosis and treatment efficacy determination. Clin Chim Acta 488:13–19. https://doi.org/10.1016/j.cca.2018.10.017
Luo X, Zhang X, Gan L, Zhou C, Zhao T, Zeng T, Liu S, Xiao Y, Yu J, Zhao F (2018) The outer membrane protein Tp92 of Treponema pallidum induces human mononuclear cell death and IL-8 secretion. J Cell Mol Med 22(12):6039–6054. https://doi.org/10.1111/jcmm.13879
Mehlitz A, Karunakaran K, Herweg JA, Krohne G, van de Linde S, Rieck E, Sauer M, Rudel T (2014) The chlamydial organism simkania negevensis forms ER vacuole contact sites and inhibits ER-stress. Cell Microbiol 16(8):1224–1243. https://doi.org/10.1111/cmi.12278
Mennerich D, Kellokumpu S, Kietzmann T (2019) Hypoxia and reactive oxygen species as modulators of endoplasmic reticulum and golgi homeostasis. Antioxid Redox Signal 30(1):113–137. https://doi.org/10.1089/ars.2018.7523
Moon HW, Han HG, Jeon YJ (2018) Protein quality control in the endoplasmic reticulum and cancer. Int J Mol Sci 19(10). https://doi.org/10.3390/ijms19103020
Moretti J, Blander JM (2017) Cell-autonomous stress responses in innate immunity. J Leukoc Biol 101(1):77–86. https://doi.org/10.1189/jlb.2MR0416-201R
Muriach M, Flores-Bellver M, Romero FJ, Barcia JM (2014) Diabetes and the brain: oxidative stress, inflammation, and autophagy. Oxidative Med Cell Longev 2014:102158. https://doi.org/10.1155/2014/102158
Peeling RW, Mabey D, Kamb ML, Chen XS, Radolf JD, Benzaken AS (2017) Syphilis. Nat Rev Dis Primers 3:17073. https://doi.org/10.1038/nrdp.2017.73
Petrosova H, Zobanikova M, Cejkova D, Mikalova L, Pospisilova P, Strouhal M, Chen L, Qin X, Muzny DM, Weinstock GM, Smajs D (2012) Whole genome sequence of Treponema pallidum ssp. pallidum strain Mexico A, suggests recombination between yaws and syphilis strains. PLoS Negl Trop Dis 6(9):e1832. https://doi.org/10.1371/journal.pntd.0001832
Rashid HO, Yadav RK, Kim HR, Chae HJ (2015) ER stress: autophagy induction, inhibition and selection. Autophagy 11(11):1956–1977. https://doi.org/10.1080/15548627.2015.1091141
Runina AV, Katunin GL, Filippova MA, Zatevalov AM, Kubanov AA, Deryabin DG (2018) Immunochip for syphilis serodiagnostics with the use of extended array of Treponema pallidum recombinant antigens. Bull Exp Biol Med 165(6):767–771. https://doi.org/10.1007/s10517-018-4261-0
Staudova B, Strouhal M, Zobanikova M, Cejkova D, Fulton LL, Chen L, Giacani L, Centurion-Lara A, Bruisten SM, Sodergren E, Weinstock GM, Smajs D (2014) Whole genome sequence of the Treponema pallidum subsp. endemicum strain Bosnia A: the genome is related to yaws treponemes but contains few loci similar to syphilis treponemes. PLoS Negl Trop Dis 8(11):e3261. https://doi.org/10.1371/journal.pntd.0003261
Tam AB, Mercado EL, Hoffmann A, Niwa M (2012) ER stress activates NF-κB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS One 7(10):e45078. https://doi.org/10.1371/journal.pone.0045078
Tu BP, Weissman JS (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164(3):341–346. https://doi.org/10.1083/jcb.200311055
Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287(5453):664–666. https://doi.org/10.1126/science.287.5453.664
Walenna NF, Kurihara Y, Chou B, Ishii K, Soejima T, Hiromatsu K (2020) Chlamydia pneumoniae infection-induced endoplasmic reticulum stress causes fatty acid-binding protein 4 secretion in murine adipocytes. J Biol Chem. https://doi.org/10.1074/jbc.RA119.010683
Weinstock GM, Hardham JM, McLeod MP, Sodergren EJ, Norris SJ (1998) The genome of Treponema pallidum: new light on the agent of syphilis. FEMS Microbiol Rev 22(4):323–332. https://doi.org/10.1111/j.1574-6976.1998.tb00373.x
Zeeshan HM, Lee GH, Kim HR, Chae HJ (2016) Endoplasmic reticulum stress and associated ROS. Int J Mol Sci 17(3):327. https://doi.org/10.3390/ijms17030327
Zhang K, Kaufman RJ (2008) From endoplasmic-reticulum stress to the inflammatory response. Nature 454(7203):455–462. https://doi.org/10.1038/nature07203
Acknowledgments
Thanks to everyone who helped us in the past 3 years.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81201331) and Major special projects of the Hunan Provincial Health and Family Planning Commission (A2017011).
Author information
Authors and Affiliations
Contributions
Performed the experiments and wrote the paper: Wei Li and Xiangping Zhou. Revised the paper: Jialun Cai, Feijun Zhao, Ting Cao, Lichang Ning, and Chunyi Luo. Critically revised the manuscript for important intellectual content: Xinhua Xiao and Shuangquan Liu. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 282 kb)
Rights and permissions
About this article
Cite this article
Li, W., Zhou, X., Cai, J. et al. Recombinant Treponema pallidum protein Tp0768 promotes proinflammatory cytokine secretion of macrophages through ER stress and ROS/NF-κB pathway. Appl Microbiol Biotechnol 105, 353–366 (2021). https://doi.org/10.1007/s00253-020-11018-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-11018-8