Abstract
Gut microbiota have been implicated in the development of cancer. Colorectal and gastric cancers, the major gastrointestinal tract cancers, are closely connected with the gut microbiome. Nevertheless, the characteristics of gut microbiota composition that correlate with gastric cancer are unclear. In this study, we investigated gut microbiota alterations during the progression of gastric cancer to identify the most relevant taxa associated with gastric cancer and evaluated the potential of the microbiome as an indicator for the diagnosis of gastric cancer. Compared with the healthy group, gut microbiota composition and diversity shifted in patients with gastric cancer. Different bacteria were used to design a random forest model, which provided an area under the curve value of 0.91. Verification samples achieved a true positive rate of 0.83 in gastric cancer. Principal component analysis showed that gastritis shares some microbiome characteristics of gastric cancer. Chemotherapy reduced the elevated bacteria levels in gastric cancer by more than half. More importantly, we found that the genera Lactobacillus and Megasphaera were associated with gastric cancer.
Key Points
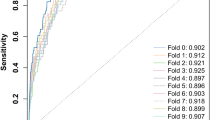
• Gut microbiota has high sensitivity and specificity in distinguishing patients with gastric cancer from healthy individuals, indicating that gut microbiota is a potential noninvasive tool for the diagnosis of gastric cancer.
• Gastritis shares some microbiota features with gastric cancer, and chemotherapy reduces the microbial abundance and diversity in gastric cancer patients.
• Two bacterial taxa, namely, Lactobacillus and Megasphaera, are predictive markers for gastric cancer.





Similar content being viewed by others
Data availability
Sequences that support the findings of this study have been deposited on NCBI under BioProject ID PRJNA639644 (available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA639644).
References
Ai D, Pan H, Li X, Wu M, Xia LC (2019) Association network analysis identifies enzymatic components of gut microbiota that significantly differ between colorectal cancer patients and healthy controls. PeerJ 7:e7315. https://doi.org/10.7717/peerj.7315
Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM (2017) Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol 14(6):356–365. https://doi.org/10.1038/nrgastro.2017.20
Alhinai EA, Walton GE, Commane DM (2019) The role of the gut microbiota in colorectal cancer causation. Int J Mol Sci 20(21). https://doi.org/10.3390/ijms20215295
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/A:1010933404324
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Castano-Rodriguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM (2013) The role of TLR2, TLR4 and CD14 genetic polymorphisms in gastric carcinogenesis: a case-control study and meta-analysis. PLoS One 8(4):e60327. https://doi.org/10.1371/journal.pone.0060327
Castano-Rodriguez N, Kaakoush NO, Pardo AL, Goh KL, Fock KM, Mitchell HM (2014) Genetic polymorphisms in the Toll-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer. Hum Immunol 75(8):808–815. https://doi.org/10.1016/j.humimm.2014.06.001
Castano-Rodriguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO (2017) Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep 7(1):15957. https://doi.org/10.1038/s41598-017-16289-2
Chen H, Jiang W (2014) Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol 5(508). https://doi.org/10.3389/fmicb.2014.00508
Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, Li Q, Wu J, Fu X (2017) Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget 8(19):31802–31814. https://doi.org/10.18632/oncotarget.15992
Chen B-y, Jiang L-x, Hao K, Wang L, Wang Y, Xie Y-w, Shen J, Zhu M-h, Tong X-m, Li K-q, Wang Z (2018) Protection of plasma transfusion against lipopolysaccharide/d-galactosamine-induced fulminant hepatic failure through inhibiting apoptosis of hepatic cells in mice. J Zhejiang Univ Sci B 19(6):436–444. https://doi.org/10.1631/jzus.B1700277
Doorakkers E, Lagergren J, Engstrand L, Brusselaers N (2016) Eradication of Helicobacter pylori and gastric cancer: a systematic review and meta-analysis of cohort studies. J Natl Cancer Inst 108(9):djw132. https://doi.org/10.1093/jnci/djw132
Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S (2001) Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol 51(Pt 4):1521–1528. https://doi.org/10.1099/00207713-51-4-1521
Farhana L, Banerjee HN, Verma M, Majumdar APN (2018) Role of microbiome in carcinogenesis process and epigenetic regulation of colorectal cancer. Methods Mol Biol 1856:35–55. https://doi.org/10.1007/978-1-4939-8751-1_3
Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, O'Riordain M, Shanahan F, O'Toole PW (2017) Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 66(4):633–643. https://doi.org/10.1136/gutjnl-2015-309595
Frick IM, Karlsson C, Morgelin M, Olin AI, Janjusevic R, Hammarstrom C, Holst E, de Chateau M, Bjorck L (2008) Identification of a novel protein promoting the colonization and survival of Finegoldia magna, a bacterial commensal and opportunistic pathogen. Mol Microbiol 70(3):695–708. https://doi.org/10.1111/j.1365-2958.2008.06439.x
Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312(5778):1355–1359. https://doi.org/10.1126/science.1124234
Hale VL, Chen J, Johnson S, Harrington SC, Yab TC, Smyrk TC, Nelson H, Boardman LA, Druliner BR, Levin TR, Rex DK, Ahnen DJ, Lance P, Ahlquist DA, Chia N (2017) Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidem Biomar 26(1):85–94. https://doi.org/10.1158/1055-9965.EPI-16-0337
Herrera V, Parsonnet J (2009) Helicobacter pylori and gastric adenocarcinoma. Clin Microbiol Infect 15(11):971–976. https://doi.org/10.1111/j.1469-0691.2009.03031.x
Holmes E, Li JV, Marchesi JR, Nicholson JK (2012) Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab 16(5):559–564. https://doi.org/10.1016/j.cmet.2012.10.007
Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, Deng YF, Hsu WT, Wu CS, Li C (2018) Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in taiwan. Sci Rep 8(1):158. https://doi.org/10.1038/s41598-017-18596-0
Jahani-Sherafat S, Alebouyeh M, Moghim S, Ahmadi Amoli H, Ghasemian-Safaei H (2018) Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol Hepatol Bed Bench 11(2):101–109
Kamada N, Seo SU, Chen GY, Nunez G (2013) Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 13(5):321–335. https://doi.org/10.1038/nri3430
Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB (2007) Dual-association of gnotobiotic IL-10-/- mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis 13(12):1457–1466. https://doi.org/10.1002/ibd.20246
Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, Zheng S, Chan FKL, Sung JJY, Yu J (2017) Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 23(8):2061–2070. https://doi.org/10.1158/1078-0432.CCR-16-1599
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb 71(12):8228–8235. https://doi.org/10.1128/aem.71.12.8228-8235.2005
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microb 73(5):1576–1585. https://doi.org/10.1128/aem.01996-06
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
McArdle BH, Anderson MJ (2001) Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82(1):290–297. https://doi.org/10.2307/2680104
Meng C, Bai C, Brown TD, Hood LE, Tian Q (2018) Human gut microbiota and gastrointestinal cancer. Genom Proteom Bioinf 16(1):33–49. https://doi.org/10.1016/j.gpb.2017.06.002
Montassier E, Batard E, Massart S, Gastinne T, Carton T, Caillon J, Le Fresne S, Caroff N, Hardouin JB, Moreau P, Potel G, Le Vacon F, de La Cochetiere MF (2014) 16S rRNA gene pyrosequencing reveals shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb Ecol 67(3):690–699. https://doi.org/10.1007/s00248-013-0355-4
Montassier E, Gastinne T, Vangay P, Al-Ghalith GA, Bruley des Varannes S, Massart S, Moreau P, Potel G, de La Cochetiere MF, Batard E, Knights D (2015) Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharmacol Ther 42(5):515–528. https://doi.org/10.1111/apt.13302
Moon JY, Zolnik CP, Wang Z, Qiu Y, Usyk M, Wang T, Kizer JR, Landay AL, Kurland IJ, Anastos K, Kaplan RC, Burk RD, Qi Q (2018) Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. EBioMedicine 37:392–400. https://doi.org/10.1016/j.ebiom.2018.10.037
Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJ (2015) Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 6:8727. https://doi.org/10.1038/ncomms9727
Nomura T, Ohkusa T, Okayasu I, Yoshida T, Sakamoto M, Hayashi H, Benno Y, Hirai S, Hojo M, Kobayashi O, Terai T, Miwa H, Takei Y, Ogihara T, Sato N (2005) Mucosa-associated bacteria in ulcerative colitis before and after antibiotic combination therapy. Aliment Pharmacol Ther 21(8):1017–1027. https://doi.org/10.1111/j.1365-2036.2005.02428.x
Ohtani N (2015) Microbiome and cancer. Semin Immunopathol 37(1):65–72. https://doi.org/10.1007/s00281-014-0457-1
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara RB, Simpson G, Solymos P, Henry M, Stevens H (2014) vegan: Community Ecology Package. (Available: https://CRAN.R-project.org/package=vegan)
Pang H, Lin A, Holford M, Enerson BE, Lu B, Lawton MP, Floyd E, Zhao H (2006) Pathway analysis using random forests classification and regression. Bioinformatics 22(16):2028–2036. https://doi.org/10.1093/bioinformatics/btl344
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30(21):3123–3124. https://doi.org/10.1093/bioinformatics/btu494
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(Database issue):D590–D596. https://doi.org/10.1093/nar/gks1219
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62(2):142–160. https://doi.org/10.1111/j.1574-6941.2007.00375.x
Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Chen X, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S (2018) Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 68:1014–1023. https://doi.org/10.1136/gutjnl-2017-315084
Rognes T, Flouri T, Nichols B, Quince C, Mahe F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J (2011) Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141(5):1782–1791. https://doi.org/10.1053/j.gastro.2011.06.072
Scott AJ, Alexander JL, Merrifield CA, Cunningham D, Jobin C, Brown R, Alverdy J, O'Keefe SJ, Gaskins HR, Teare J, Yu J, Hughes DJ, Verstraelen H, Burton J, O'Toole PW, Rosenberg DW, Marchesi JR, Kinross JM (2019) International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 68(9):1624–1632. https://doi.org/10.1136/gutjnl-2019-318556
Song M, Chan A, Sun J (2020) Influence of the Gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology 158(2):322–340. https://doi.org/10.1053/j.gastro.2019.06.048
Thorell K, Bengtsson-Palme J, Liu OH-F, Palacios Gonzales RV, Nookaew I, Rabeneck L, Paszat L, Graham DY, Nielsen J, Lundin SB, Sjöling Å (2017) In vivo analysis of the viable microbiota and Helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infect Immun 85(10):e00031–e00017. https://doi.org/10.1128/IAI.00031-17
Vinasco K, Mitchell HM, Kaakoush NO, Castano-Rodriguez N (2019) Microbial carcinogenesis: lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer 1872(2):188309. https://doi.org/10.1016/j.bbcan.2019.07.004
Vineis JH, Ringus DL, Morrison HG, Delmont TO, Dalal S, Raffals LH, Antonopoulos DA, Rubin DT, Eren AM, Chang EB, Sogin ML (2016) Patient-specific Bacteroides genome variants in pouchitis. MBio 7(6). https://doi.org/10.1128/mBio.01713-16
Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L (2012) Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6(2):320–329. https://doi.org/10.1038/ismej.2011.109
Wang LL, Yu XJ, Zhan SH, Jia SJ, Tian ZB, Dong QJ (2014) Participation of microbiota in the development of gastric cancer. World J Gastroenterol 20(17):4948–4952. https://doi.org/10.3748/wjg.v20.i17.4948
Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, Dong Q (2016) Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol 28(3):261–266. https://doi.org/10.1097/MEG.0000000000000542
Wang YW, Du YQ, Miao XL, Ye GY, Wang YY, Xu AB, Jing YZ, Tong Y, Xu K, Zheng MQ, Chen D, Wang Z (2018) Risk factors and drug resistance in early-onset neonatal group B streptococcal disease. J Zhejiang Univ Sci B 19(12):973–978. https://doi.org/10.1631/jzus.B1800165
Wong SH, Kwong TNY, Wu CY, Yu J (2018) Clinical applications of gut microbiota in cancer biology. Semin Cancer Biol 55:28–36. https://doi.org/10.1016/j.semcancer.2018.05.003
Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, Wang L, Wang C, Ding T, Ravel J, Taylor PR, Abnet CC, Goldstein AM (2017) Molecular characterization of the human stomach microbiota in gastric cancer patients. Front Cell Infect Mi 7(302). https://doi.org/10.3389/fcimb.2017.00302
Acknowledgments
We thank Mingyi Wo (Zhejiang Provincial People’s Hospital) for the help in sample collection and thank Yong Wang (Hangzhou Guhe Information and Technology Company) for the assistance in the data analysis. We would like to acknowledge the participants for providing samples.
Funding
This work was supported by the Medicine and Health Research Foundation of Zhejiang Province in China (2019RC012, 2019KY017, 2017KY216 and 2017KY486), and Key projects jointly constructed by the Ministry and the province of Zhejiang Medical and Health Science and Technology Project in China (WKJ-ZJ-2019). Key and Major Projects of Traditional Chinese Medicine Scientific Research Foundation of Zhejiang Province in China (2019ZZ001).
Author information
Authors and Affiliations
Contributions
JXL and ZW designed the study. YYZ and JS contributed to writing the manuscript. YYZ, GLJ, and YFN provided technical and material support and data analysis; XWS and YQD provided the samples and clinical data.
Corresponding authors
Ethics declarations
Competing interests
Yaofang Niu and Gulei Jin from the Hangzhou Guhe Information and Technology Company. We declare no potential conflicts of interest were disclosed by the other authors.
Ethics approval
This work has been improved by the ethics committee of Zhejiang Provincial People’s Hospital.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 910 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Shen, J., Shi, X. et al. Gut microbiome analysis as a predictive marker for the gastric cancer patients. Appl Microbiol Biotechnol 105, 803–814 (2021). https://doi.org/10.1007/s00253-020-11043-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-11043-7