Abstract
Staphylococcus aureus is a serious pathogen unleashing its virulence through several classes of exotoxins such as hemolysins and enterotoxins. In this study, we designed a novel multi-antigen subunit vaccine which can induce innate, humoral and cellular immune responses. Alpha hemolysin, enterotoxins A and B were selected as protective antigens for combining into a triple antigen chimeric protein (HAB). Immunoinformatics analysis predicted HAB protein as a suitable vaccine candidate for inducing both humoral and cellular immune responses. Tertiary structure of the HAB protein was predicted and validated through computational approaches. Docking studies were performed between the HAB protein and mice TLR2 receptor. Furthermore, we constructed and generated recombinant HAB (r-HAB) protein in E. coli and studied its toxicity, immunogenicity and protective efficacy in a mouse model. Triple antigen chimeric protein (r-HAB) was found to be highly immunogenic in mouse as the anti-r-HAB hyperimmune serum was strongly reactive to all three native exotoxins on Western blot. In vitro toxin neutralization assay using anti-r-HAB antibodies demonstrated > 75% neutralization of toxins on RAW 264.7 cell line. Active immunization with r-HAB toxoid gave ~ 83% protection against 2 × lethal dosage of secreted exotoxins. The protection was mediated by induction of strong antibody responses that neutralized the toxins. Passive immunization with anti-r-HAB antibodies gave ~ 50% protection from lethal challenge. In conclusion, in vitro and in vivo testing of r-HAB found the molecule to be nontoxic, highly immunogenic and induced excellent protection towards native toxins in actively immunized and partial protection to passively immunized mice groups.
Key points
• HAB protein was computationally designed to induce humoral and cellular responses.
• r-HAB protein was found to be nontoxic, immunogenic and protective in mouse model.
• r-HAB conferred protection against lethal challenge in active and passive immunization.
Similar content being viewed by others
Introduction
Staphylococcus aureus is a Gram-positive, common resident microflora on humans and a potential opportunistic pathogen (Kota et al. 2020). At least 70–80% of healthy adult population will be carriers of S. aureus either permanently or intermittently (Zhao et al. 2021). Carriers are asymptomatic in most cases; however, those individuals with immunological or barrier defects and preceding other viral or bacterial infections are at high risk of acquiring invasive infections (Wertheim et al. 2005). Despite several attempts to develop protective vaccines and immunotherapies, neither approach was effective in preventing recurring staphylococcal infections in humans (Fowler and Proctor 2014). Difficulties in developing prophylactic and therapeutic measures against S. aureus emerge from the fact that, organism possesses multiple arsenals of virulence and immune evasion factors, multiple manifestations of infections and our limited understanding of the intricacies of host–pathogen interactions (Fowler and Proctor 2014; Reddy et al. 2017). Lack of protective licensed vaccine combined with dramatic increase of antibiotic-resistant strains in hospital and community at alarming rate has further added to treatment woes (Klausberger et al. 2019).
Vaccine targeting S. aureus should comprise multiple components each contributing to protective efficacy against some manifestation of infection. Exotoxins produced by S. aureus play a crucial role in pathogenesis, invasiveness and disease severity (Reddy et al. 2017; Hemmadi and Biswas 2021). Hemolysins and enterotoxins are different classes of exotoxins that have prominent role in several staphylococcal diseases. Alpha hemolysin is a pore forming exotoxin expressed by most S. aureus strains and a direct correlation between the amount of alpha hemolysin expression and virulence of the strain was established suggesting its prominent role in pathogenesis (Wardenburg and Schneewind 2008). Enterotoxin A (SEA) and enterotoxin B (SEB) belong to class of heat stable pyrogenic toxin superantigens (PTSA) secreted by certain S. aureus strains and are implicated in food poisoning and toxic shock syndrome. SEA is most common and potent enterotoxin responsible for food intoxication. SEB is a powerful exotoxin implicated in food intoxication and is also a potent inducer of T-cells and hence it has been listed as a category B biological warfare agent (Reddy et al. 2015). Several variants of short, truncated or mutated versions of these toxins were used to induce protective antibody responses against the native toxins (Kennedy et al. 2010; Adhikari et al. 2012, 2016; Uppalapati et al. 2012; Reddy et al. 2015). Lessons from previous preclinical and clinical research suggest that an effective vaccine should comprise multiple antigens targeting different classes of virulence factors (Begier et al. 2017).
In the present study, a multi-antigen subunit vaccine (HAB) carrying truncated and inactive forms of alpha hemolysin and enterotoxin A and B was designed comprising B-cell and T-cell epitopes which can induce robust humoral and cellular immune responses. Both B-cell and T-cell epitopes in the HAB protein were predicted through in silico methods. Immunoinformatics approaches were employed for structural analysis of the vaccine candidate. Immunoinformatics enables researchers to identify and design suitable vaccine candidates against bacterial pathogens such as S. aureus (Afley et al. 2015; Kolla et al. 2021). Physicochemical analysis, 3-D structural analysis and molecular docking with TLR2 receptor were also carried out during the study. The recombinant version of HAB (r-HAB) protein was experimentally produced in E. coli strain by constructing r-hab fusion gene by using overlap extension PCR and evaluated its immunogenicity, safety, tolerability and protective efficacy in murine model. The ability of r-HAB protein in inducing humoral and toxin neutralization responses was evaluated by in vitro and in vivo methods such as active immunization, antibody titre determination, toxin neutralization, passive immunotherapy and lethal challenge studies. Alpha hemolysin, enterotoxin A and enterotoxin B were chosen to be included based on their toxicity, frequency of occurrence and catering different families of exotoxins.
Materials and methods
Materials
Dehydrated microbial culture media, antibiotics and supplements are procured from HiMedia laboratories, India. pET22b vector, Escherichia coli NovaBlue and BL21(DE3) pLysS strains were from Novagen, USA. Mouse monoclonal anti-6x-histidine antibodies and HRP-conjugated secondary antibodies were from Sigma-Aldrich, India. Inorganic chemicals and organic solvents were from Sisco Research Laboratory, India.
Antigen selection and in silico cloning
Nucleotide and amino acid sequences of alpha hemolysin (Hla), staphylococcal enterotoxin A and enterotoxin B (SEA and SEB) were retrieved from National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) bearing accession numbers X01645.1, M18970.1 and M11118.1 respectively. Partial portions of the three exotoxins previously proven to be protective were screened for their antigenicity (Adhikari et al. 2016; Reddy et al. 2015; Shylaja et al. 2012). Antigenicity of the three peptides was determined with VaxiJen 2.0 server at default threshold value 0.4 (http://www.ddgpharmfac.net/vaxijen/VaxiJen/VaxiJen.html) (Doytchinova and Flower 2008). Furthermore, the gene sequences of the three antigens were cloned in to pET22b vector to generate triple antigen chimeric protein HAB employing GeneRunner software without any alteration in the open reading frames. A flexible Gly4Ser (G4S) linker was placed in between hla and sea genes. Furthermore, seb gene was cloned adjacent to sea gene separated by AAGCTTCTCGAG (KLLE) spacer sequence. Nucleotide sequence of HAB was translated into corresponding amino acid sequence for computational analysis.
Immunogenicity, stability and physicochemical attributes
The fusion protein HAB was further analysed for its immunological and biochemical properties. Antigenicity of the protein was determined with VaxiJen 2.0 server at default threshold 0.4 (http://www.ddgpharmfac.net/vaxijen/VaxiJen/VaxiJen.html). VaxiJen 2.0 is an alignment independent server based on auto-cross covariance transformation of protein sequences (Doytchinova and Flower 2008). Physicochemical parameters such as theoretical pI, molar extinction coefficient, instability index, aliphatic index and Grand average of hydropathicity (GRAVY) were determined with ProtParam resource (https://web.expasy.org/protparam/) (Gasteiger et al. 2005). ProtParam is an online tool available at Swiss bioinformatics institute. The homology of the developed antigen was checked by using amino acid sequence of the HAB protein and subjecting for protein BLAST search against the proteomes of higher animals such as Mus musculus (mice), Cavia porcellus (guinea pig), Rattus (rat), Bos taurus (bovine) and Homo sapiens (Humans).
Tertiary structure prediction and validation
Three-dimensional tertiary structure of HAB protein was predicted to study its interaction with toll-like receptors during innate immune response. Tertiary structure of the protein was predicted with RaptorX server (http://raptorx.uchicago.edu/). RaptorX is a distance-based protein folding algorithm developed through deep learning (Källberg et al. 2014). Model generated by the RaptorX server was refined prior to structural analysis. Structural refinement was performed by GalaxyRefine server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE). This server refines the protein structure through Ab-initio loop modelling (Ko et al. 2012). Structural quality of the generated model was assessed based on its Z-score and extent of falling of model in to the characteristic range of protein structures determined through X-ray crystallography and NMR spectroscopic methods. Z-score of the predicted protein structure was estimated with ProSA-web server (Wiederstein and Sippl 2007).
Epitope prediction
B-cell and T-cell epitopes in HAB protein were determined through in silico methods. Linear B-cell epitopes were predicted with BepiPred-2.0 server (http://www.cbs.dtu.dk/services/BepiPred/) (Jespersen et al. 2017). Discontinuous epitopes in HAB protein were predicted with DiscoTope tool (http://tools.iedb.org/discotope/). Discotope predicts the discontinuous epitopes in the vaccine molecule based on folding pattern in the tertiary structure of the protein (Anderson et al. 2006). Cytotoxic T-cell epitopes were predicted against H-2-Dd, H-2-Kd and H-2-Ld MHC I alleles of BALB/c mice (Zhang et al. 2005) with NetCTLpan 1.1 server (http://www.cbs.dtu.dk/services/NetCTLpan/) (Stranzl et al. 2010). T-helper cell epitopes against BALB/c mice MHC II alleles H-2-IAd, H-2-IEd (Zhang et al. 2005) were predicted with NetMHCIIpan 4.0 server (Reynisson et al. 2020).
Molecular docking
Molecular docking is a computational simulation program that mimics the biological process in life systems. Molecular docking studies were conducted on HAB protein and mice Toll-like receptor 2 (TLR2). TLR2 is involved in innate response as a part of anti-microbial defence mechanisms. Three-dimensional crystal structure of mice TLR2 was downloaded from RCSB-PDB (PDB ID-3A79) (https://www.rcsb.org/). Native ligands and water molecules were removed from the protein crystal structure using PyMol software package. Molecular docking between chain A of HAB antigen and chain B of TLR2 was performed with GRAMM-X server. GRAMM-X server employs refinement and scoring functions for protein docking and simulations (Tovchigrechko and Vakser 2006). Gibbs free energy between ligand and receptor in the docked complex was calculated with PRODIGY portal in the HADDOCK web server (https://wenmr.science.uu.nl/prodigy/) (Xue et al. 2016; Branco and Choupina 2021). Accessible surface area in both the peptide chains and contact map of hydrophobic, hydrophilic and hydrogen bonds between the two peptide chains in the docked complex was visualized with CoCoMaps tool (https://www.molnac.unisa.it/BioTools/cocomaps/view.psp#asa1) (Vangone et al. 2011).
Bacterial strains and toxin preparation
Staphylococcus aureus ATCC 29213 strain was used for gene amplification by PCR and for toxin production. S. aureus ATCC 29213 strain is positive for alpha hemolysin, enterotoxin B and enterotoxin A production. For gene amplification by PCR, genomic DNA from this strain was isolated by phenol:chloroform:isoamyl alcohol (25:24:1) extraction after lysostaphin (Sigma-Aldrich) treatment to digest the cell wall (Gill 2013). For toxin production, fresh overnight culture (5 mL) grown in Tryptic Soy broth (TSB) was inoculated in fresh 250 mL of TSB and grown at 37 °C for 24 h with vigorous shaking at 200 RPM. Cells were separated by centrifugation and cell free supernatant (CFS) was filtered through 0.22-μm syringe filter (Pall life Sciences, India). Filtered culture supernatants were subjected to methanol/chloroform precipitation and protein concentrate was dissolved in 1/20th volume of phosphate buffered saline (PBS). Concentrated protein preparation was subjected to dialysis against 1 × PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4 pH 7.4) to get rid of media components. Dialysis was performed against minimum of 1000 sample volumes of 1 × PBS buffer. PBS buffer was changed 4 times during 24-h dialysis period.
Determination of lethal dose for crude toxin preparation
Different volumes (10–200 μl) of concentrated crude toxin preparation from S. aureus ATCC 29,213 containing all three exotoxins (Hla, SEA, SEB) were initially tested on naïve mice to determine the lethal dose (LD) before undertaking challenge studies. Exotoxin preparation was administered to mice through intraperitoneal route and the condition of mice was observed on the following day. Least quantity of toxin preparation which killed mice was considered as the lethal dose. For challenge studies, 2 × quantity of lethal dose was used for both active and passively immunized mice groups.
Primer designing
Primers used for amplification of alpha hemolysin, entertoxin A and exterotoxin B are given in Table 1. Oligonucleotide pairs were designed for amplification of N-terminal region of alpha hemolysin. Amplified hla gene portion does not contain signal sequence and includes key residues for oligomerization. Similarly, conserved regions of enterotoxin A and B were amplified using the primers listed in Table 1. The amino acid identity of SEA and SEB with other SEs ranged from 20 to 50% (Table 2).
PCR amplification, gene fusion and cloning
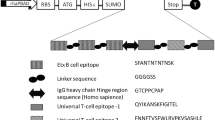
Gene fragments of hla, sea and seb were PCR amplified using oligonucleotides listed in Table 1. Primers carried restriction enzyme recognition sequences or linker sequences at their 5′ ends. Alpha hemolysin and enterotoxin A genes were fused using overlap extension PCR with a central flexible glycine-serine linker (G4S) and cloned in BamHI and HindIII sites of pET22b vector. Recombinant plasmid (pET22b-hla-sea) carrying alpha hemolysin and enterotoxin A genes was extracted followed by cloning the enterotoxin B gene fragment adjacent to enterotoxin A fragment in XhoI restriction site. Schematic representation of the cloning strategy is given in Fig. 1. The recombinant plasmid carrying triple toxin fusion was transformed in chemically competent E. coli NovaBlue strain by heat shock at 42 °C. The plasmid extracted from recombinant E. coli NovaBlue was further transformed into expression host E. coli BL21(DE3) pLysS (Novagen, USA).
Expression and purification
Overnight culture (2 mL) of E. coli BL21(DE3) pLysS carrying pET22b-hla-sea-seb recombinant plasmid was added to 200 mL of Luria–Bertani broth containing 100 μg/mL ampicillin and 35 μg/mL chloramphenicol. The culture was incubated at 37 °C with vigorous shaking at 200 RPM. Once the culture O.D600 reached ~ 0.6, the triple toxin chimeric gene (hla-sea-seb) cloned in pET22b under the control of T7 promoter was induced by adding 1 mM of isopropyl β-D-galactopyranoside (IPTG). The culture was incubated for another 5 h, after which the cells were harvested by centrifugation at 9000 g. Histidine (6 × his) tagged recombinant protein was purified under native conditions from periplasmic extracts using imidazole and Ni–NTA resin by immobilized metal affinity chromatography (IMAC) (Qiagen, Germany). Eluents carrying recombinant protein were pooled, buffer exchanged in 1 × PBS, lyophilized and resuspended in sterile distilled water. Concentration of purified protein was quantified by Lowry’s colorimetric assay using BSA as standard (Lowry et al. 1951).
Safety studies, immunization and antibody titre testing
At the outset, safety of r-HAB multi-antigen vaccine was studied in 5–6-week-old female BALB/c mice and observed for potential side effects such as pain, discomfort, fever, emesis, hemolysis and deaths after administering 2 × times the immunization dosage through intraperitoneal route without adjuvants. After confirming the safety of r-HAB, immunization of r-HAB protein was undertaken in 5–6-week-old specific pathogen-free female BALB/c mice bred in in-house animal facility (Reg. No.: 2046/PO/ReBi/S/18/CPCSEA) of Vignan’s University. Three groups of 6 mice each were separated and housed in pairs belonging to same immunization groups. Approximately 50-μg quantity of recombinant protein was mixed and emulsified with equal volume of Freund’s complete adjuvant (FCA) and delivered subcutaneously to group 1 mice. Same group received two more booster doses (30 μg) in Freund’s incomplete adjuvant (FIA) at 14-day interval through subcutaneous route. Group 2 mice received PBS emulsified with FCA and FIA through subcutaneous route. Group 3 mice were given only sterile 1 × PBS through intraperitoneal route. Mice blood was sampled 2 days before start of immunization and 12 days past each immunization and the antibody titres were determined by indirect ELISA (Engvall and Perlmann 1972; Kota et al. 2020) using recombinant r-HAB as antigen. Least dilution showing absorbance twice the value of PBS only group of mice is considered the end-point titres.
Western blot analysis
Western blot analysis was carried out to check the reactivity of anti-HAB serum against all the three native exotoxins from S. aureus cultures. S. aureus strains grown for 24–36 h in tryptic soy broth at 37 °C were centrifuged and the supernatants containing native exotoxins were concentrated 10-folds by methanol:chloroform precipitation method. Concentrated culture supernatants (CCS) were separated on 12% SDS-PAGE gels in tris–glycine buffer. Separated proteins were transferred on to nitrocellulose membrane by wet transfer in Bio-Rad mini transblot apparatus. Transferred blots were incubated overnight at 4 °C in 5% non-fat skim milk prepared in 1 × PBS. Blots were washed with 1 × PBST (tween — 0.05%) to get rid of excess and unbound milk proteins. Later blots were incubated with 1:2000 dilutions of anti-r-HAB hyperimmune serum and incubated at room temperature (RT) for 45 min. Blots were treated with PBST followed by addition of secondary goat anti-mouse antibody conjugated to horseradish peroxidase (HRP). Blots were washed 4–5 times in PBST followed by development with diaminobenzidine tetrahydrochloride (DAB) (Sigma-Aldrich, India) and 0.03% H2O2 in PBS.
Antibody isotype analysis
Blood collected 12 days after the last immunization from different mice groups was used for determining antibody isotypes (Reddy et al. 2015). Antibody isotypes in hyperimmune serum was quantified using mouse antibody isotyping reagents (Sigma-Aldrich). Serum was tested for IgG1, IgG2a, IgG2b, IgG3, IgM and IgA antibodies. Control and test sera were diluted to 1:100 dilutions in PBS and 0.1 mL was coated on to the wells of Nunc microtitre plates followed by isotype determination by indirect ELISA as per manufacturer’s instructions. Appearance of deep orange colour indicated positive reaction for appropriate isotype. The absorbance was measured at 490 nm in Multiskan sky microplate spectrophotometer (Thermo, USA).
Toxin neutralization assay
Toxin neutralization potential of polyclonal anti-HAB antibodies was assessed on RAW 264.7 cell line. Briefly, crude toxins isolated and concentrated from 24 h S. aureus cultures were treated with different dilutions of antisera (1:100, 1:250, 1:500). The effect of toxins on cell viability and morphology was evaluated and the ability of serum antibodies to neutralize the toxins was established by MTT assay and microscopic studies. MTT assay was performed in 96-well cell culture plates (Nunc) (Reddy et al. 2015). Briefly, cells were cultured at 37 °C with 5% CO2 until ~ 70% confluency. Later cells were incubated with toxin and different serum dilutions in DMEM and incubated for 24 h. Serum from control group (PBS) of mice served as negative control. Spent media were aspirated and 50 μL of 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrasolium bromide (MTT) in DMEM was added to the wells and incubated for further 4 h at 37 °C. DMSO was added to solubilize the formazan crystals. Absorbance was measured at 570 nm. Toxin neutralization percentage was calculated using the formula: (OD of toxin only) – (OD of toxin + antibody)/(OD of toxin only) – (OD of no toxin) × 100. The morphology of the treated and untreated cells was observed using phase contrast microscopy (Olympus).
Animal challenge studies
Protective efficacy of r-HAB protein against native exotoxins was evaluated in BALB/c mouse. For active challenge, 2 × lethal dose of toxin preparation was injected intraperitoneally into the immunized group (6 no’s) of mice 10 days after the third dosage of antigen administration. In case of passive challenge group (6 no’s), antiserum (250 μl) collected from actively immunized mice was administered intravenously and left for 24 h. Passively immunized mice were challenged with 2 × lethal dose of exotoxin preparation. A control group (6 no’s) of mice receiving only adjuvants were also challenged with 2 × lethal dose of exotoxin preparation. Challenged mice groups were observed for 10 days post-challenge and any deaths occurred were recorded for calculating the survival percentage.
Statistical analysis
All statistical analyses were performed in GraphPad Prism 8. ELISA and antibody titre testing were performed in three repetitions and expressed as mean ± standard deviation (SD). For antibody titre testing, statistical difference between experimental groups was compared using univariate ANOVA and the difference at P < 0.05 was considered to be significant. For antibody isotypes, unpaired t-test was applied. Kaplan–Meier survival graphs were constructed in GraphPad Prism 8.
Results
Immunogenicity, stability and physicochemical properties
Antigenicity of the HAB protein was predicted as 0.5956. Similarly, antigenic scores of the three peptides, viz. alpha-hemolysin, SEA and SEB were 0.6412, 0.5441 and 0.5132 respectively. Biochemical parameters such as theoretical pI, molar extinction coefficient, instability index, aliphatic index and grand average of hydropathicity (GRAVY) of HAB protein are 6.44, 72,910 M−1 cm−1, 32.70, 67.27 and − 0.830 respectively. GRAVY value indicates the protein as hydrophilic. Proteins with aliphatic index range 66.5–84.33 and instability index less than 40 are stable at in vitro conditions. Stability of the protein at in vitro conditions was crucial for its biological activity. Furthermore, no significant match for the antigen was observed against the proteomes of higher animals during BLASTP analysis. Hence, we presume that r-HAB antigen cannot induce auto-immune reactions in the higher animals including humans. Thus, HAB protein was computationally evaluated to be stable at in vitro conditions. Similarly, antigenic proteins are very crucial for protective immune response. Therefore, the HAB was found to be a better subunit vaccine.
Structural analysis
Raptor-X server generated 5 models of r-HAB protein. Model 1 with estimated RMSD 9.4093 Å was considered for further analysis. This structure was further refined with GalaxyWEB server. Structural refinement aims to model protein structure with high accuracy. GalaxyWEB generated 5 refined models from the predicted protein structure. Model 1 out of the 5 models has 96.1% of residues in the favoured region of Ramachandran plot. Z-score of this model was − 8.65 indicating the characteristic range of X-ray crystallographic structure and does not include any errors in the structure. Therefore, this model was found to be energetically stable and structurally reliable for further studies (Fig. 2a). Structural analysis led to the identification of immunodominant regions in the antigen because the efficacious and protective immune response requires the identification of certain exposed regions in the antigen structure.
(a) Three-dimensional tertiary structure of HAB triple antigen chimeric protein predicted with RaptorX server from the deduced amino acid sequence. Truncated regions of alpha-hemolysin, SEA and SEB are represented in red, cyan and blue colours. G4S linker sequence that joins alpha-hemolysin and SEA antigens was highlighted in green colour. Yellow coloured domains indicate the vector parts. Hexa-histidine tag was coloured with hot pink. (b) Docked complex between mice TLR2 receptor and HAB protein ligand. Pink coloured chain represents TLR2 receptor. Green coloured macromolecule represents the HAB protein ligand. (c) Contact map of interactions between TLR2 receptor and HAB ligand. Pink, yellow and green colour dots specify the hydrophilic-hydrophilic, hydrophilic-hydrophobic and hydrophobic-hydrophobic interactions between the chain A and chain B in the docked complex. Amino acid residues of mice TLR-2 and HAB protein are represented by vertical and horizontal axes. (d) Conformational B-cell epitopes in the antigen. Regions highlighted in yellow colour are the antibody binding domains or conformational B-cell epitopes in the HAB antigen
Molecular docking
Molecular docking studies were performed between mice TLR2 and HAB protein with GRAMM-X server. TLR2 is involved in anti-microbial defence mechanisms against S. aureus infections during innate immune response. GRAMM-X server generated 10 docked models. Models generated by GRAMM-X server were ordered by scoring functional value of the model. Best model among the generated docked complexes was ranked first and placed top in the output file by the server (Tovchigrechko and Vakser 2006). Model 1 was selected as best one from the output file (Fig. 2b). Gibbs free energy (ΔG) between HAB protein and mice TLR2 was − 17.4 kcalMol−1. Therefore, a strong interaction between HAB and TLR2 was established in the docked complex. Contact of docked complex was visualized to identify the type of interactions between HAB antigen and TLR2 that are involved in activating innate immune response. Totally 83 residues from TLR2 and 78 residues from HAB protein were involved in interaction between the two chains (Fig. 2c). Forty-eight residues from TLR2 and 39 residues from HAB protein are present in the accessible surface area (ASA) regions. Amino acids 1–150 of TLR2 with 250–350 amino acids of HAB and 550–575 amino acids of TLR2 with 1–100 amino acids of HAB were identified as interaction islands in the docked complex. Two hundred and five hydrophilic-hydrophobic interactions, 173 hydrophilic-hydrophilic interactions, 60 hydrophobic-hydrophobic interactions and 6 hydrogen bonds are present in docked complex ensuring high affinity and strong interaction between TLR2 and HAB protein.
Epitope mapping
B-cell and T-cell epitopes in the HAB protein are listed in Table 3. BepiPred-2.0 predicted 7 linear B-cell epitopes in HAB protein. Two linear B-cell epitopes each were identified in alpha hemolysin and SEA portions. Three linear epitopes were found in SEB antigen (Table 3). NetCTLpan and NetMHCIIpan predicted cytotoxic T-cell and T-helper cell epitopes in HAB protein (Table 3). Alpha-hemolysin comprised both cytotoxic T-cell and T-helper cell epitopes, whereas only T-helper cell epitopes were present on both the enterotoxins SEA and SEB portions. Among all the cytotoxic T-cell epitopes, WPSAFKVQL, MHKKVFYSF, TIAGQYRVY and NTTVKTGDL are found to be frequently presented from the r-HAB through all the MHC alleles in BALB/c mice during antigen processing and presentation in innate immune response. Structural and discontinuous epitopes in HAB protein were predicted with DiscoTope server of IEDB (Immune epitope database). A total of 237 residues were found in antibody binding domains in HAB protein (regions highlighted in yellow colour in the protein structure). Both the receptor bound and secretory immunoglobulins of B-cells bind to these regions of HAB protein during humoral immune response (Fig. 2d).
Gene fusion and cloning
Partial hla gene (297 bp) coding for the key residues required for oligomerization and hemolytic activity was amplified by PCR. Similarly, PCR amplified sea and seb genes were 366 and 666 bp respectively in length and comprised conserved portions of staphylococcal enterotoxins (Fig. 5a). Partial hla and sea genes were combined by gene fusion and further cloned in pET22b vector. In the resulting recombinant vector (pET22b-hla-seb), the seb gene was cloned by restriction and ligation procedure. The combined fusion gene is under the control of T7 promoter. Hla and SEA portions were separated by a single repeat of glycine – serine linker (G4S). Gene coding for SEB was cloned later in the recombinant pET22b-hla-sea plasmid. The total length of fusion gene after cloning was 1369 bp by nested PCR with outer hla forward and seb reverse primers (Fig. 3a). Recombinant fusion gene was sequenced using T7 primers and the resultant DNA sequence was in complete agreement with the expected sequence.
PCR amplification, gene fusion, cloning and expression of r-HAB protein. Representation of immunization, blood and serum sampling, antibody and isotypes determination. (a) PCR amplification of partial genes and cloning. Lanes 1, 2 and 3 represent partial sea, seb and hla genes. Lane 4 represents the complete fusion gene (sea-seb-hla) after cloning PCR amplified from pET-22b-hla-sea-seb plasmid by colony PCR. (b) Coomassie brilliant blue stained SDS-PAGE gel showing expressed r-HAB protein after IPTG induction. I — induced with IPTG; UI — uninduced. Panel (c) Blot treated with anti-His antibodies showing expressed r-HAB protein after IPTG induction. I — induced with IPTG; UI — uninduced. (d) Schematic representation of immunization, blood sampling, antibody titre determination and blood collection schedules. EG — experimental group; CG — control group; FCA — Freund’s complete adjuvant; FIA — Freund’s incomplete adjuvant. Panel (e) Estimation of antibody titre against r-HAB antigen at different points after immunization. (f) Antibody isotypes determination for IgG subtypes, IgA and IgM antibodies. Data are represented in mean ± SD (n = 8). Significance (P)-value summary analysed by univariate ANOVA: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; #no significance
Expression and purification of triple fusion protein
Induction of triple fusion gene with IPTG resulted in expression of protein at 56-kDa region as evidenced by in silico cloning using GeneRunner 6.5, SDS-PAGE staining and Western blot analysis (Fig. 3b and 3c). The chimeric r-HAB protein was expressed with a carboxy terminal 6 × histidine tag which is part of the vector. Recombinant protein was found localized in periplasmic fraction and hence it was purified under native conditions using imidazole. The yield of the recombinant protein was ~ 6 mg/100-mL culture. Recombinant protein was subjected to buffer exchange with sterile 1 × PBS by dialysis. Recombinant protein was purified under native conditions in order to retain the structural features of native proteins to the maximum extent possible for optimal immune response.
Safety studies, immunization and antibody titre testing
None of the mice displayed any visible discomfort, isolated behaviour, pain or emesis. All the mice survived the 30-day observation period. We presume that the toxicity associated with individual natural toxins was eradicated when the partial portions were combined in to a fusion molecule. Antigen immunization and blood collection schedules are shown in Fig. 3d. Recombinant HAB protein induced strong humoral responses in all the individual mice as seen in progressive increase of antibody titres with each successive immunization. Figure 3e represents the antibody titres of six numbers of immunized group of mice. The results indicated the immunogenic nature of the recombinant antigen. Antibody titres were tested on days − 2, 12, 26 and 40 for 0th, 1st, 2nd and 3rd immunization doses respectively. Antibody titres reached 1:64,000 dilutions after the 3rd immunization when tested by indirect ELISA with r-HAB as antigen. Sera from the control group of mice did not exhibit reactivity to the recombinant antigen. On 40th day, blood collection was carried out and sera was pooled from immunized and control group of mice for further experimental analysis.
Isotype analysis
Isotype-specific monoclonal antibodies were used to evaluate anti-r-HAB serum isotypes by ELISA. Isotyping analysis revealed predominance of IgG1 and IgG2b isotypes when compared to IgG2a, IgG3, IgM and IgA isotypes and subtypes. A Th2-type immune response was elicited by r-HAB antigen as seen from predominance of IgG1 antibody. Furthermore, even the presence of IgG2b antibodies at moderate levels indicated a Th1-type polarized humoral immune responses. Levels of IgM, IgA and IgG2a isotypes were comparable to control mice serum. Recombinant HAB toxoid in conjunction with Freund’s adjuvant induced primarily Th-2 type of humoral responses in mice model as the IgG1:IgG2b ratio was observed to be > 1 (Fig. 3f).
Western blot analysis
Western blot analysis revealed the immune reactivity of anti-r-HAB serum towards all the three native exotoxins — alpha hemolysin, enterotoxin A and enterotoxin B. Partial and truncated portions of the exotoxins as a single fusion molecule were able to induce strong humoral responses in mice as seen from high antibody titres and reactivity to native toxins on blots. Immune reactivity to native toxins confirmed that the periplasmic localization and buffer exchange of the r-HAB protein helped preserve epitopes identical to the native toxin. Reactivity to alpha hemolysin was observed at ~ 33-kDa region, SEA and SEB toxins at 28-kDa region on the blots (Fig. 4). Blots treated with control serum did not exhibit any reactivity at the expected regions. Spurious and unrelated bands seen on the blots at 50–60-kDa region indicate the cross-reactivity of staphylococcal protein A — a natural Ig binding protein present in the culture supernatants to primary and/or secondary antibodies.
Determination of lethal dose for crude toxin preparation
Methanol/chloroform concentration procedure retained the biological activity of exotoxin preparation. Approximately 150-μl quantity of crude exotoxin preparation was determined to be the lethal dose for undertaking challenge studies. In this study, crude toxin preparation was preferred over purified toxins in order to reflect the real S. aureus infection scenario. For assessing the protective efficacy of r-HAB toxoid and anti-r-HAB serum in active and passive immunization procedures respectively, 2 × quantity (300 μl) of exotoxin preparation was used for challenging the mice.
Toxin neutralization assay
Anti-r-HAB serum at 1:100 dilution was able to partially neutralize the toxic effect of exotoxins on the RAW 264.7 cells. Cells were protected from lysis and death when exposed to the exotoxin preparation preincubated with the anti-r-HAB serum antibodies from immunized mice. MTT assay was chosen to calculate the percentage of metabolically active cells upon incubation with toxin and antibodies. At 1/100 dilution of serum antibodies, ~ 78% of toxin neutralization was recorded based on absorbance of RAW 264.7 cells in the presence of toxin preincubated with antibodies. Cells treated with toxin plus 1/100 dilution of antibody in comparison with control cells were nearly identical in cell morphology, ultrastructure, cell count and metabolic activity (Fig. 5a). Crude toxins collected from culture supernatants of pure cultures were used for neutralization studies to mimic a real S. aureus infection scenario. Due to the polyclonal nature of the serum and multiple isotypes of antibodies, the neutralization was observed to be more pronounced.
(a) Phase contrast images of RAW 264.7 cells under different treatment conditions. a1. Control cells without any treatment. a2. Cells treated with concentrated exotoxin preparation. a3. Cell treated with exotoxin preparation preincubated with r-HAB antiserum. Cells were rescued by the neutralizing antibodies present in the serum. (b) Kaplan–Meier survival curves showing percentage survival in different mice groups. Mice group receiving r-HAB antigen showed 83% survival rate when compared to 0% survival in control groups. Similarly, passively immunized mice group showed 50% survival rate compared to 0% in control groups
Animal challenge studies
Toxoid nature of r-HAB protein was evaluated through active and passive immunization studies. Mice were kept for observation for 10 days for possible deaths. In case of actively immunized mice (n = 6) administered with 2 × lethal dose of toxin preparation, only one animal succumbed to death and other animals were protected from toxaemia and death (83% survival). In case of the control mice (n = 6) administered with same quantity of exotoxin preparation, all the mice died within 24 h. Similarly, another batch of control group of mice (n = 6) were passively immunized with 250-μl quantity of serum by intravenous route followed by exposure to 2 × dose of exotoxin preparation. Approximately, 50% of mice (n = 3) were protected from hemolysin and superantigen induced toxicity and survived beyond the observation period of 10 days. Mice which succumbed to death in case of passively challenged mice died before 96 h (4 days) (Fig. 5b).
Discussion
Development of novel antibiotics is expensive and complicated than development of subunit vaccines and therapeutic antibodies. Furthermore, there is always a risk of bacteria acquiring the resistance against antibiotics. Most humans live in frequent contact with S. aureus and therefore the natural colonized individuals have low titres of anti-S. aureus antibodies in them. However, S. aureus is naturally equipped with multiple factors to disarm the host antibodies. Therefore, the goal of vaccines and immunotherapies is to equip individual more protection than already available in most patients. Earlier strategies of vaccine development for S. aureus were aimed at promoting opsonophagocytic bacterial uptake and subsequent killing. These efforts have failed to deliver a safe and protective vaccine. Understanding the effect of S. aureus and its virulence determinants on host adaptive immunity during infection will help to identify promising vaccine targets. Several epidemiological and experimental studies have stressed that S. aureus exotoxins are prominent drivers of pathogen virulence. Therefore, toxoids and neutralizing antibodies represent a viable approach to preventing or reducing the mortality and morbidity of S. aureus infections (Kailasan et al. 2019). S. aureus produces multiple classes of exotoxins such as hemolysins, enterotoxins, leukocidins, phenol soluble modulins (PSMs), proteases and protease modulators causing varied manifestations of disease (Singh and Phukan 2019). Virulence of S. aureus strain is attributed to the number of exotoxins being possessed by that strain. Thus, targeting multiple virulence factors tackling different disease manifestations is suggested for vaccine, toxoid design or for passive antibody therapy. S. aureus alpha hemolysin is a 33-kDa heptameric pore forming cytotoxin responsible for dermonecrosis, myositis, pneumoniae, necrotizing fasciitis and mastitis (Uppalapati et al. 2014). The heptameric form of the cytotoxin binds to membranes of several cell types and form pores and also induce arachidonic acid metabolism leading to vasoconstriction, cell death and tissue necrosis (Dinges et al. 2000). Studies have proved that virulence of a S. aureus strain is clearly linked to the presence and amount of alpha toxin secreted (Bubeck Wardenburg et al. 2008; Reddy et al. 2017). Staphylococcal enterotoxins (SEs) comprise more than 20 toxins belonging to pyrogenic toxin superantigen (PTSA) superfamily and are structurally and functionally related, thermostable and secretory exotoxins which induce emetic activity (He et al. 2019; Pinchuk et al. 2010; Reddy et al. 2015). SEA and SEB together constitute the reason for majority of the staphylococcal food poisoning cases worldwide. Both SEA and SEB are powerful T-cell activators responsible for over activating the immune system and leading to multiple organ failure and death.
In recent years, numerous advancements in in silico tools and algorithms, statistical methods and enormous growth of numerous databases have expanded the sequence-based methodologies in vaccine research and development (Hajialibeigi et al. 2021). Therefore, in this study, a recombinant chimeric triple antigen toxoid r-HAB was designed, developed and found to be highly immunogenic molecule able to induce strong immune response against the native toxins. Epitope mapping has shown that the antigen can induce both cellular and humoral immune responses. Nearly 50% of the residues are present in antibody binding domains in the antigen. This was reflected in in vitro assay using RAW 264.7 mammalian cell line where r-HAB antibodies protected the cells from toxin effects when the toxins were preincubated with serum antibodies. Molecular docking has the ability to predict the binding-conformation between ligand and protein and the amino acid residues that are being responsible for the interaction which cannot be determined through conventional experimental studies (Vakser 2014). A strong interaction was discovered between the antigen and mice TLR2 during molecular docking studies. This represents the interaction between TLR2 and antigen during innate immune response. Consequently, we observed that mice groups receiving the experimental r-HAB protein were > 80% protective during toxin challenge and partially protective during passive challenge in a mouse model. These results were consistent with other works where in S. aureus, exotoxins generate excellent humoral responses which bring neutralization of toxin components (Kailasan et al. 2019; Uppalapati et al. 2012). These results were similar to our earlier study where we reported the construction of a fusion between enterotoxin A and toxic shock syndrome toxin (TSST-1) which showed protection against the lethal dose of crude SEA and TSST-1 toxins (Reddy et al. 2015). Furthermore, portions of SEA and SEB in r-HAB shared 25–50% homology with other staphylococcal enterotoxins. We assume that the r-HAB antibodies might show similar or partial neutralization effects to other SEs. Neutralizing antibodies aid in preventing toxinosis thus reducing the severity of S. aureus infections. Recombinant HAB toxoid was highly potent and induced antibodies which were reactive to native toxins. In active immunized mice, r-HAB conferred excellent protection to lethal toxin challenge. However, with passively immunized mice, the protection conferred was partial.
This study highlights the importance of inclusion of exotoxins as vaccine antigens against S. aureus. Alpha hemolysin-based toxoid immunization protected the host from cytotoxic and necrotic effects. Similarly, enterotoxin-based toxoid protected the host from immune overactivation and possible toxic shock syndrome. Thus, immunity against cytolytic and superantigens not only inhibits their biological activity but protects the host from their effects. The protective effects of the r-HAB are limited to strains harbouring alpha hemolysin and enterotoxins A and B. Therefore, more antigens need to be included to address the strains that harbour other or no toxins to achieve protection against whole pathogen. We hypothesize that inclusion of a surface molecule such as lipoteichoic acid (LTA) and another protein entity such as clumping factor (clfA) and fibronectin binding proteins (fnbpA/ fnbpB) might be efficacious (Narita et al. 2017). Ultimately a successful vaccine must elicit both humoral and T-cell responses that will neutralize the pathogen through promoting bacterial clearance and restricting immune evasion.
Data availability
All data generated or analysed in this study are included in this article.
This article describes the study involving BALB/c mice for which necessary permission was sought from Institutional Animal Ethical Committee (IAEC) of Vignan’s Foundation for Science, Technology and Research. The protocols and procedures performed in this study abide the guidelines laid by the Committee for the purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India, for proper care and use of experimental animals.
References
Adhikari RP, Karauzum H, Sarwar J, Abaandou L, Mahmoudieh M, Boroun AR, Vu H, Nguyen T, Devi VS, Shulenin S, Warfield KL, Aman MJ (2012) Novel structurally designed vaccine for S. aureus α-hemolysin: protection against bacteremia and pneumonia. PloS one 7(6):e38567. https://doi.org/10.1371/journal.pone.0038567
Adhikari RP, Thompson CD, Aman MJ, Lee JC (2016) Protective efficacy of a novel alpha hemolysin subunit vaccine (AT62) against Staphylococcus aureus skin and soft tissue infections. Vaccine 34(50):6402–6407. https://doi.org/10.1016/j.vaccine.2016.09.061
Afley P, Dohre SK, Prasad GBKS, Kumar S (2015) Prediction of T cell epitopes of Brucella abortus and evaluation of their protective role in mice. Appl Microbiol Biotechnol 99(18):7625–7637. https://doi.org/10.1007/s00253-015-6787-7
Andersen PH, Nielsen M, Lund O (2006) Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci 15(11):2558–2567. https://doi.org/10.1110/ps.062405906
Begier E, Seiden DJ, Patton M, Zito E, Severs J, Cooper D, Eiden J, Gruber WC, Jansen KU, Anderson AS, Gurtman A (2017) SA4Ag, a 4-antigen Staphylococcus aureus vaccine, rapidly induces high levels of bacteria-killing antibodies. Vaccine 35(8):1132–1139. https://doi.org/10.1016/j.vaccine.2017.01.024
Branco I, Choupina A (2021) Bioinformatics: new tools and applications in life science and personalized medicine. Appl Microbiol Biotechnol 105:937–951. https://doi.org/10.1007/s00253-020-11056-2
BubeckWardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR (2008) Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis 198(8):1166–1170. https://doi.org/10.1086/592053
Dinges MM, Orwin PM, Schlievert PM (2000) Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 13:16–34. https://doi.org/10.1128/CMR.13.1.16
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 8:4. https://doi.org/10.1186/1471-2105-8-4
Engvall E, Perlmann P (1972) Enzyme-linked immunosorbent assay Elisa. 3. Quantitation of specific antibodies by enzyme-labeled anti-immunoglobulin in antigen-coated tubes. J Immunol 109(1):129–135
Fowler VG Jr, Proctor RA (2014) Where does a Staphylococcus aureus vaccine stand? Clin Microbiol Infect 20:66–75. https://doi.org/10.1111/1469-0691.12570
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. Humana press, New Jersey, pp 571–607. https://doi.org/10.1385/1-59259-890-0:571
Gill. (2013). S. aureus genomic DNA isolation. OpenWetWare. https://openwetware.org/wiki/Gill:S._aureus_genomic_DNA_isolation
Hajialibeigi A, Amani J, Gargari SLM (2021) Identification and evaluation of novel vaccine candidates against Shigella flexneri through reverse vaccinology approach. Appl Microbiol Biotechnol 105(3):1159–1173
He Y, Sun Y, Ren Y, Qiao L, Guo R, Du J, Zhu X, Liu Y, Lin J (2019) The T cell activating properties and antitumour activity of Staphylococcal Enterotoxin-like Q. Med Microbiol Immunol 208(6):781–792. https://doi.org/10.1007/s00430-019-00614-9
Hemmadi V, Biswas M (2021) An overview of moonlighting proteins in Staphylococcus aureus infection. Arch Microbiol 203:481–498. https://doi.org/10.1007/s00203-020-02071-y
Jespersen MC, Peters B, Nielsen M, Marcatili P (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45(W1):W24–W29. https://doi.org/10.1093/nar/gkx346
Kailasan S, Kort T, Mukherjee I, Liao GC, Kanipakala T, Williston N, Ganjbaksh N, Venkatasubramaniam A, Holtsberg FW, Karauzum H, Adhikari RP, Aman MJ (2019) Rational design of toxoid vaccine candidates for Staphylococcus aureus Leukocidin AB (LukAB). Toxins 11(6):339. https://doi.org/10.3390/toxins11060339
Källberg M, Margaryan G, Wang S, Ma J, Xu J (2014) RaptorX server: a resource for template-based protein structure modeling. Methods Mol Biol 1137:17–27. https://doi.org/10.1007/978-1-4939-0366-5_2
Kennedy AD, Wardenburg JB, Gardner DJ, Long D, Whitney AR, Braughton KR, Schneewind O, DeLeo FR (2010) Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis 202(7):1050–1058. https://doi.org/10.1086/656043
Klausberger M, Leneva IA, Falynskova IN, Vasiliev K, Poddubikov AV, Lindner C, Kartaschova NP, Svitich OA, Stukova M, GrabherrM E, A, (2019) The potential of influenza HA-specific immunity in mitigating lethality of post-influenza pneumococcal infections. Vaccines 7(4):187. https://doi.org/10.3390/vaccines7040187
Ko J, Park H, Heo L, Seok C (2012) GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res 40:W294–W297. https://doi.org/10.1093/nar/gks493
Kolla HB, Tirumalasetty C, Sreerama K (2021) Ayyagari VS (2021) An immunoinformatics approach for the design of a multi-epitope vaccine targeting super antigen TSST-1 of Staphylococcus aureus. J Genet EngBiotechnol 19:69. https://doi.org/10.1186/s43141-021-00160-z
Kota RK, Reddy PN, Sreerama K (2020) Application of IgY antibodies against staphylococcal protein A (SpA) of Staphylococcus aureus for detection and prophylactic functions. Appl Microbiol Biotechnol 104(21):9387–9398. https://doi.org/10.1007/s00253-020-10912-5
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Narita K, Asano K, Nakane A (2017) IL-17A plays an important role in protection induced by vaccination with fibronectin-binding domain of fibronectin-binding protein A against Staphylococcus aureus infection. Med Microbiol Immunol 206(3):225–234. https://doi.org/10.1007/s00430-017-0499-9
Pinchuk IV, Beswick EJ, Reyes VE (2010) Staphylococcal Enterotoxins Toxins 2(8):2177–2197. https://doi.org/10.3390/toxins2082177
Reddy PN, Paul S, Sripathy MH, Batra HV (2015) Evaluation of recombinant SEA-TSST fusion toxoid for protection against superantigen induced toxicity in mouse model. Toxicon 103:106–113. https://doi.org/10.1016/j.toxicon.2015.06.008
Reddy PN, Srirama K, Dirisala VR (2017) An update on clinical burden, diagnostic tools, and therapeutic options of Staphylococcus aureus. Infect Dis: Res Treat 10:1179916117703999. https://doi.org/10.1177/1179916117703999
Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M (2020) Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res 19(6):2304–2315. https://doi.org/10.1021/acs.jproteome.9b00874
Shylaja R, Thakasi DKK, Murali HS, Reddy KPN, Batra HV (2012) Application of a chimeric protein construct having enterotoxin B and toxic shock syndrome toxin domains of S. aureus in immunodiagnostics. Indian J Microbiol 5(3):449–455. https://doi.org/10.1007/s12088-012-0269-8
Singh V, Phukan UJ (2019) Interaction of host and Staphylococcus aureus protease-system regulates virulence and pathogenicity. Med Microbiol Immunol 208(5):585–607. https://doi.org/10.1007/s00430-018-0573-y
Stranzl T, Larsen MV, Lundegaard C, Nielsen M (2010) NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics 62(6):357–368. https://doi.org/10.1007/s00251-010-0441-4
Tovchigrechko A, Vakser IA (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res 34:W310–W314. https://doi.org/10.1093/nar/gkl206
Uppalapati SR, Kingston JJ, Murali HS, Batra HV (2012) Generation and characterization of an inter-generic bivalent alpha domain fusion protein αCS from Clostridium perfringens and Staphylococcus aureus for concurrent diagnosis and therapeutic applications. J Appl Microbiol 113(2):448–458. https://doi.org/10.1111/j.1365-2672.2012.05333.x
Uppalapati SR, Kingston JJ, Murali HS, Batra HV (2014) Heterologous protection against alpha toxins of Clostridium perfringens and Staphylococcus aureus induced by binding domain recombinant chimeric protein. Vaccine 32(25):3075–3081. https://doi.org/10.1016/j.vaccine.2014.03.021
Vakser IA (2014) Protein-protein docking: from interaction to interactome. Biophys J 107(8):1785–1793. https://doi.org/10.1016/j.bpj.2014.08.033
Vangone A, Spinelli R, Scarano V, Cavallo L, Oliva R (2011) COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics 27(20):2915–2916. https://doi.org/10.1093/bioinformatics/btr484
Wardenburg JB, Schneewind O (2008) Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med 205(2):287–294. https://doi.org/10.1084/jem.20072208
Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5(12):751–762. https://doi.org/10.1016/S1473-3099(05)70295-4
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids 35:W407–W410. https://doi.org/10.1093/nar/gkm290
Xue LC, Rodrigues JP, Kastritis PL, Bonvin AM, Vangone A (2016) PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32(23):3676–3678. https://doi.org/10.1093/bioinformatics/btw514
Zhang GL, Srinivasan KN, Veeramani A, August JT, Brusic V (2005) PREDBALB/c: a system for the prediction of peptide binding to H2d molecules, a haplotype of the BALB/c mouse. Nucleic Acids Res 33:W180–W183. https://doi.org/10.1093/nar/gki479
Zhao N, Cheng D, Jian Y, Liu Y, Liu J, Huang Q, He L, Wang H, Miao F, Li M, Liu Q (2021) Molecular characteristics of Staphylococcus aureus isolates colonizing human nares and skin. Med Micro Ecol 7:100031. https://doi.org/10.1016/j.medmic.2020.100031
Acknowledgements
Prakash Narayana Reddy thanks INSPIRE division of Department of Science and Technology (DST), Government of India (GOI), for granting INSPIRE faculty award and research grant during the study period. Authors thank Department of Science and Technology, GOI, for conferring FIST support to the Biotechnology department of Vignan’s University for 5 years (2015–2019) (File No: SR/FST/LSI-576/2013(C)). Rohini Krishna Kota thanks Council of Scientific and Industrial Research (CSIR), Govt. of India, for awarding Senior Research Fellowship (SRF) (File no.: 09/1253(0002)/2019-EMR-I). Authors thank Dr. Aravind Shekar for proofreading the manuscript and for offering critical comments.
Funding
Funding was provided by the Department of Science and Technology, Government of India, under the INSPIRE Faculty programme to Prakash Narayana Reddy (File No: DST/INSPIRE/04/2017/000565).
Author information
Authors and Affiliations
Contributions
PNR conceived and designed the research. RKK, HBK, SKS and NKK performed the experimental work. PNR and HBK wrote the manuscript. The final version of manuscript was read and approved by all the authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kota, R.K., Kolla, H.B., Reddy, P.N. et al. Immunoinformatics analysis and evaluation of recombinant chimeric triple antigen toxoid (r-HAB) against Staphylococcus aureus toxaemia in mouse model. Appl Microbiol Biotechnol 105, 8297–8311 (2021). https://doi.org/10.1007/s00253-021-11609-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11609-z