Abstract
The use of messenger RNA (mRNA) profiling is considered a promising method in the identification of forensically relevant body fluids which can provide crucial information for reconstructing a potential crime. However, casework samples are usually of limited quantity or have been subjected to degradation, which requires improvement of body fluid identification. Circular RNAs (circRNAs), a class of products from the backsplicing of pre-mRNAs, are shown to have high abundance, remarkable stability, and cell type-specific expression in human cells. In this study, we investigated whether the inclusion of circRNAs in mRNA profiling improve the detection of biomarkers including δ-aminolevulinate synthase 2 (ALAS2) and matrix metallopeptidase 7 (MMP7) in body fluid identification. The major circRNAs of ALAS2 and MMP7 were first identified and primer sets for the simultaneous detection of linear and circular transcripts were developed. The inclusion of circRNAs in mRNA profiling showed improved detection sensitivity and stability of biomarkers revealed by using serial dilutions, mixed samples, and menstrual bloodstains as well as degraded and aged samples. Therefore, the inclusion of circRNAs in mRNA profiling should facilitate the detection of mRNA markers in forensic body fluid identification.




Similar content being viewed by others
References
Virkler K, Lednev IK (2009) Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci Int 188:1–17. https://doi.org/10.1016/j.forsciint.2009.02.013
Fleming RI, Harbison S (2010) The development of a mRNA multiplex RT-PCR assay for the definitive identification of body fluids. Forensic Sci Int Genet 4:244–256. https://doi.org/10.1016/j.fsigen.2009.10.006
Richard ML, Harper KA, Craig RL, Onorato AJ, Robertson JM, Donfack J (2012) Evaluation of mRNA marker specificity for the identification of five human body fluids by capillary electrophoresis. Forensic Sci Int Genet 6:452–460. https://doi.org/10.1016/j.fsigen.2011.09.007
Park JL, Park SM, Kwon OH et al (2014) Microarray screening and qRT-PCR evaluation of microRNA markers for forensic body fluid identification. Electrophoresis 35:3062–3068. https://doi.org/10.1002/elps.201400075
Sauer E, Reinke AK, Courts C (2016) Differentiation of five body fluids from forensic samples by expression analysis of four microRNAs using quantitative PCR. Forensic Sci Int Genet 22:89–99. https://doi.org/10.1016/j.fsigen.2016.01.018
An JH, Choi A, Shin KJ, Yang WI, Lee HY (2013) DNA methylation-specific multiplex assays for body fluid identification. Int J Legal Med 127:35–43. https://doi.org/10.1007/s00414-012-0719-1
Park JL, Kwon OH, Kim JH et al (2014) Identification of body fluid-specific DNA methylation markers for use in forensic science. Forensic Sci Int Genet 13:147–153. https://doi.org/10.1016/j.fsigen.2014.07.011
Giampaoli S, Berti A, Valeriani F et al (2012) Molecular identification of vaginal fluid by microbial signature. Forensic Sci Int Genet 6:559–564. https://doi.org/10.1016/j.fsigen.2012.01.005
Choi A, Shin KJ, Yang WI, Lee HY (2014) Body fluid identification by integrated analysis of DNA methylation and body fluid-specific microbial DNA. Int J Legal Med 128:33–41. https://doi.org/10.1007/s00414-013-0918-4
Haas C, Hanson E, Anjos MJ et al (2012) RNA/DNA co-analysis from blood stains—results of a second collaborative EDNAP exercise. Forensic Sci Int Genet 6:70–80. https://doi.org/10.1016/j.fsigen.2011.02.004
Haas C, Hanson E, Anjos MJ et al (2013) RNA/DNA co-analysis from human saliva and semen stains—results of a third collaborative EDNAP exercise. Forensic Sci Int Genet 7:230–239. https://doi.org/10.1016/j.fsigen.2012.10.011
Lindenbergh A, van den Berge M, Oostra RJ et al (2013) Development of a mRNA profiling multiplex for the inference of organ tissues. Int J Legal Med 127:891–900. https://doi.org/10.1007/s00414-013-0895-7
Song F, Luo H, Hou Y (2015) Developed and evaluated a multiplex mRNA profiling system for body fluid identification in Chinese Han population. J Forensic Legal Med 35:73–80. https://doi.org/10.1016/j.jflm.2015.08.006
Lux C, Schyma C, Madea B, Courts C (2014) Identification of gunshots to the head by detection of RNA in backspatter primarily expressed in brain tissue. Forensic Sci Int 237:62–69. https://doi.org/10.1016/j.forsciint.2014.01.016
Winter J, Diederichs S (2011) Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 8:1149–1157. https://doi.org/10.4161/rna.8.6.17665
Setzer M, Juusola J, Ballantyne J (2008) Recovery and stability of RNA in vaginal swabs and blood, semen, and saliva stains. J Forensic Sci 53:296–305. https://doi.org/10.1111/j.1556-4029.2007.00652.x
Sirker M, Schneider PM, Gomes I (2016) A 17-month time course study of human RNA and DNA degradation in body fluids under dry and humid environmental conditions. Int J Legal Med 130:1431–1438. https://doi.org/10.1007/s00414-016-1373-9
Lin MH, Albani PP, Fleming R (2015) Degraded RNA transcript stable regions (StaRs) as targets for enhanced forensic RNA body fluid identification. Forensic Sci Int Genet 20:61–70. https://doi.org/10.1016/j.fsigen.2015.09.012
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. https://doi.org/10.1371/journal.pone.0030733
Jeck WR, Sorrentino JA, Wang K et al (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. https://doi.org/10.1261/rna.035667.112
Memczak S, Jens M, Elefsinioti A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:409. https://doi.org/10.1186/s13059-014-0409-z
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159:134–147. https://doi.org/10.1016/j.cell.2014.09.001
Starke S, Jost I, Rossbach O et al (2015) Exon circularization requires canonical splice signals. Cell Rep 10:103–111. https://doi.org/10.1016/j.celrep.2014.12.002
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388. https://doi.org/10.1080/15476286.2015.1020271
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777. https://doi.org/10.1371/journal.pgen.1003777
Haas C, Hanson E, Bar W et al (2011) mRNA profiling for the identification of blood—results of a collaborative EDNAP exercise. Forensic Sci Int Genet 5:21–26. https://doi.org/10.1016/j.fsigen.2010.01.003
Haas C, Hanson E, Kratzer A, Bar W, Ballantyne J (2011) Selection of highly specific and sensitive mRNA biomarkers for the identification of blood. Forensic Sci Int Genet 5:449–458. https://doi.org/10.1016/j.fsigen.2010.09.006
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. https://doi.org/10.1038/nbt.2890
Lindenbergh A, de Pagter M, Ramdayal G et al (2012) A multiplex (m)RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Sci Int Genet 6:565–577. https://doi.org/10.1016/j.fsigen.2012.01.009
Haas C, Klesser B, Maake C, Bar W, Kratzer A (2009) mRNA profiling for body fluid identification by reverse transcription endpoint PCR and realtime PCR. Forensic Sci Int Genet 3:80–88. https://doi.org/10.1016/j.fsigen.2008.11.003
Juusola J, Ballantyne J (2007) mRNA profiling for body fluid identification by multiplex quantitative RT-PCR. J Forensic Sci 52:1252–1262. https://doi.org/10.1111/j.1556-4029.2007.00550.x
Juusola J, Ballantyne J (2005) Multiplex mRNA profiling for the identification of body fluids. Forensic Sci Int 152:1–12. https://doi.org/10.1016/j.forsciint.2005.02.020
Roeder AD, Haas C (2013) mRNA profiling using a minimum of five mRNA markers per body fluid and a novel scoring method for body fluid identification. Int J Legal Med 127:707–721. https://doi.org/10.1007/s00414-012-0794-3
van den Berge M, Bhoelai B, Harteveld J, Matai A, Sijen T (2015) Advancing forensic RNA typing: on non-target secretions, a nasal mucosa marker, a differential co-extraction protocol and the sensitivity of DNA and RNA profiling. Forensic Sci Int Genet 20:119–129. https://doi.org/10.1016/j.fsigen.2015.10.011
Ashwal-Fluss R, Meyer M, Pamudurti NR et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Houseley J, Tollervey D (2009) The many pathways of RNA degradation. Cell 136:763–776. https://doi.org/10.1016/j.cell.2009.01.019
Acknowledgements
The authors thank Dr. Xiao-Ou Zhang from the Chinese Academy of Sciences in Shanghai for his help with the analysis of circRNAs. This work was supported by the National Natural Science Foundation of China (81571853 and 31270862).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Figure S1
The strategy for the detection of linear and/ or circular transcripts. (A) Outward-facing primer sets (orange arrows) were designed based on exons shared by linear and circular transcripts. As for outward-facing primer sets, the binding sites of forward primers (F) in the coding sequence are located downstream from the binding sites of reverse primers (R), which result in the failure of PCR amplification on linear transcripts. Therefore, outward-facing primer sets were used for the detection of circRNAs in this study. (B) Conventional primer sets were designed based on at least one exon employed only by linear transcripts (L-primers, red arrows) for the detection of linear transcripts. Conventional primer sets were designed based on exons shared by linear and circular transcripts (LC-primers, green arrows) for the detection of linear and circular transcripts. F: forward primers; R: reverse primers. (GIF 13 kb)
Figure S2
(A) One putative circRNAs of HBA identified by bioinformatics analysis is indicated by the triangle and one circular transcript identified in (C) is indicated by the asterisk. (B and C) Detection of circRNAs of ALAS2 (B) and HBA (C) in the peripheral blood using outward-facing primer sets for PCR amplification followed by AGE. (D) The conjunct sequence of the circular HBA transcript identified in (C). The arrow indicates the junction site. (E) A longer conjunct sequence of the ALAS2 circRNA identified in (B) and the arrow indicates the junction site. (F) Detection of circRNAs of MMP7 in the menstrual blood using outward-facing primer sets for PCR amplification followed by AGE. (GIF 83 kb)
Figure S3
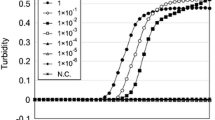
(A) Dissociation curve from the real-time PCR assay using L-primers (left) and LC-primers (right) of ALAS2. (B) Determination of the amplification efficiencies using LC-primers of ALAS2 (left) as well as L-primers (middle) and LC-primers (right) of MMP-7 by qPCR. The panels indicate the mathematical relationships of the Ct value and the copy number of full-length DNA. Each point represents the mean of three replicates. (GIF 43 kb)
Figure S5
(A, B) The RFU values from the detection of ALAS2 using L-primers and LC-primers in three 13 day-old (A) and 18 day-old (B) peripheral bloodstains were calculated. (C, D) The RFU values from the detection of MMP7 using L-primers and LC-primers in three 13 day-old (C) and 18 day-old (D) menstrual bloodstains were calculated. (GIF 41 kb)
Table S1
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Liu, B., Shao, C. et al. Evaluation of the inclusion of circular RNAs in mRNA profiling in forensic body fluid identification. Int J Legal Med 132, 43–52 (2018). https://doi.org/10.1007/s00414-017-1690-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-017-1690-7