Abstract
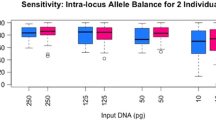
Analyzing genetic markers in nuclear and mitochondrial genomes is helpful in various forensic applications, such as individual identifications and kinship analyses. However, most commercial kits detect these markers separately, which is time-consuming, laborious, and more error-prone (mislabelling, contamination, ...). The MGIEasy Signature Identification Library Prep Kit (hereinafter “MGIEasy identification system”; MGI Tech, Shenzhen, China) has been designed to provide a simple, fast, and robust way to detect appropriate markers in one multiplex PCR reaction: 52 autosomal STRs, 27 X-chromosomal STRs, 48 Y-chromosomal STRs, 145 identity-informative SNPs, 53 ancestry-informative SNPs, 29 phenotype-informative SNPs, and the hypervariable regions of mitochondrial DNA (mtDNA). Here, we validated the performance of MGIEasy identification system following the guidelines of the Scientific Working Group on DNA Analysis Methods (SWGDAM), assessing species specificity, sensitivity, mixture identification, stability under non-optimal conditions (degraded samples, inhibitor contamination, and various substrates), repeatability, and concordance. Libraries prepared using MGIEasy identification system were sequenced on a MGISEQ-2000 instrument (MGI Tech). MGIEasy-derived STR, SNP, and mtDNA genotypes were highly concordant with CE-based STR genotypes (99.79%), MiSeq FGx-based SNP genotypes (99.78%), and Sanger-based mtDNA genotypes (100%), respectively. This system was strongly human-specific, resistant to four common PCR inhibitors, and reliably amplified both low quantities of DNA (as low as 0.125 ng) and degraded DNA (~ 150 nt). Most of the unique alleles from the minor contributor were detected in 1:10 male-female and male-male mixtures; some minor Y-STR alleles were even detected in 1:1000 male-female mixtures. MGIEasy also successfully directly amplified markers from blood stains on FTA cards, filter papers, and swabs. Thus, our results demonstrated that MGIEasy identification system was suitable for use in forensic analyses due to its robust and reliable performance on samples of varying quality and quantity.









Similar content being viewed by others
References
Liu YY, Harbison SA (2018) A review of bioinformatic methods for forensic DNA analyses. Forensic Sci Int Genet 33:117–128
Butler JM, Coble MD, Vallone PM (2007) STRs vs. SNPs: thoughts on the future of forensic DNA testing. Forensic Sci Med Pathol 3:200–205
Tamaki K, Kaszynski RH, Yuan QH, Yoshida K, Okuno T, Tsuruyama T (2009) Likelihood evaluation using 15 common short tandem repeat loci: a practical and simulated approach to establishing personal identification via sibling/parental assessments. Transfusion 49:578–584
Poetsch M, Lüdcke C, Repenning A, Fischer L, Mályusz V, Simeoni E, Lignitz E, Oehmichen M, von Wurmb-Schwark N (2006) The problem of single parent/child paternity analysis-practical results involving 336 children and 348 unrelated men. Forensic Sci Int 159:98–103
Aceves MEG, Cortés GM, Villalobos HR (2017) Results obtained in five years in a paternity testing laboratory in Mexico. Forensic Sci Int Genet Suppl Ser 6:e305–e307
Canturk KM, Emre R, Kínoglu K et al (2014) Current status of the use of single-nucleotide polymorphisms in forensic practices. Genet Test Mol Biomark 18:455–460
Wenk RE (2004) Testing for parentage and kinship. Curr Opin Hematol 11:357–361
Amorim A, Pereira L (2005) Pros and cons in the use of SNPs in forensic kinship investigation: a comparative analysis with STRs. Forensic Sci Int 150:17–21
Tamura T, Osawa M, Ochiai E, Suzuki T, Nakamura T (2015) Evaluation of advanced multiplex short tandem repeat systems in pairwise kinship analysis. Legal Med 17:320–325
Fridman C, Gonzalez RS, Pereira AC, Cardena MMSG (2014) Haplotype diversity in mitochondrial DNA hypervariable region in a population of southeastern Brazil. Int J Legal Med 128:589–593
Holland MM, Parsons TJ (1999) Mitochondrial DNA sequence analysis - validation and use for forensic casework. Forensic Sci Rev 11:21–50
Hao WQ, Liu J, Jiang L, Han JP, Wang L, Li JL, Ma Q, Liu C, Wang HJ, Li CX (2019) Exploring the ancestry differentiation and inference capacity of the 28-plex AISNPs. Int J Legal Med 133:975–982
Arnold C (2020) The controversial company using DNA to sketch the faces of criminals. Nature 585:178–181
Zhou H, Bi G, Zhang C, Liu Y, Chen R, Li F, Mei X, Guo Y, Zheng W (2016) Developmental validation of forensic DNA-STR kits: expressmarker 16 + 10Y and expressmarker 16 + 18Y. Forensic Sci Int Genet 24:1–17
Xie M, Song F, Li J, Xie B, Wang S, Wang W, Ma H, Luo H (2020) Validation of the AGCU Expressmarker 16 + 22Y Kit: a new multiplex for forensic application. Int J Legal Med 134:177–183
Jäger AC, Alvarez ML, Davis CP, Guzmán E, Han Y, Way L, Walichiewicz P, Silva D, Pham N, Caves G, Bruand J, Schlesinger F, Pond SJK, Varlaro J, Stephens KM, Holt CL (2017) Developmental validation of the MiSeq FGx forensic genomics system for targeted next generation sequencing in forensic DNA casework and database laboratories. Forensic Sci Int Genet 28:52–70
Børsting C, Morling N (2015) Next generation sequencing and its applications in forensic genetics. Forensic Sci Int Genet 18:78–89
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351
Jeon SA, Park JL, Kim JH, Kim JH, Kim YS, Kim JC, Kim SY (2019) Comparison of the MGISEQ-2000 and Illumina Hiseq 4000 sequencing platforms for RNA sequencing. Genomics Inform 17(3):e32
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352
de la Puente M, Phillips C, Santos C, Fondevila M, Carracedo Á, Lareu MV (2017) Evaluation of the Qiagen 140-SNP forensic identification multiplex for massively parallel sequencing. Forensic Sci Int Genet 28:35–43
Woerner AE, Ambers A, Wendt FR, King JL, Moura-Neto RS, Silva R, Budowle B (2018) Evaluation of the precision ID mtDNA whole genome panel on two massively parallel sequencing systems. Forensic Sci Int Genet 36:213–224
Pereira V, Mogensen HS, Børsting C, Morling N (2017) Evaluation of the Precision ID Ancestry Panel for crime case work: a SNP typing assay developed for typing of 165 ancestral informative markers. Forensic Sci Int Genet 28:138–145
Strobl C, Eduardoff M, Bus MM, Allen M, Parson W (2018) Evaluation of the precision ID whole MtDNA genome panel for forensic analyses. Forensic Sci Int Genet 35:21–25
Meiklejohn KA, Robertson JM (2017) Evaluation of the Precision ID Identity Panel for the Ion Torrent™ PGM™ sequencer. Forensic Sci Int Genet 31:48–56
Mo SK, Ren ZL, Yang YR, Liu YC, Zhang JJ, Wu HJ, Li Z, Bo XC, Wang SQ, Yan JW, Ni M (2018) A 472-SNP panel for pairwise kinship testing of second-degree relatives. Forensic Sci Int Genet 34:178–185
Liu Q, Ma G, Du Q et al (2020) Development of an NGS panel containing 42 autosomal STR loci and the evaluation focusing on secondary kinship analysis. Int J Legal Med 134(6):2005–2014
Tao R, Qi W, Chen C, Zhang J, Yang Z, Song W, Zhang S, Li C (2019) Pilot study for forensic evaluations of the Precision ID GlobalFiler™ NGS STR Panel v2 with the Ion S5™ system. Forensic Sci Int Genet 43:102147
Natarajan KN, Miao Z, Jiang M, Huang X, Zhou H, Xie J, Wang C, Qin S, Zhao Z, Wu L, Yang N, Li B, Hou Y, Liu S, Teichmann SA (2019) Comparative analysis of sequencing technologies for single-cell transcriptomics. Genome Biol 20(1):70
Fehlmann T, Reinheimer S, Geng C, Su X, Drmanac S, Alexeev A, Zhang C, Backes C, Ludwig N, Hart M, An D, Zhu Z, Xu C, Chen A, Ni M, Liu J, Li Y, Poulter M, Li Y, Stähler C, Drmanac R, Xu X, Meese E, Keller A (2016) cPAS-based sequencing on the BGISEQ-500 to explore small non-coding RNAs. Clin Epigenetics 8:123
Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, Qu S, Mei X, Chen H, Yu T, Sun N, Rao J, Wang J, Zhang W, Chen Y, Liao S, Jiang H, Liu X, Yang Z, Mu F, Gao S (2017) A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience 6:1–9
Mak SST, Gopalakrishnan S, Carøe C, Geng C, Liu S, Sinding MS, Kuderna LFK, Zhang W, Fu S, Vieira FG, Germonpré M, Bocherens H, Fedorov S, Petersen B, Sicheritz-Pontén T, Marques-Bonet T, Zhang G, Jiang H, Gilbert MTP (2017) Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience 6(8):1–13
Fang C, Zhong H, Lin Y, Chen B, Han M, Ren H, Lu H, Luber JM, Xia M, Li W, Stein S, Xu X, Zhang W, Drmanac R, Wang J, Yang H, Hammarström L, Kostic AD, Kristiansen K, Li J (2018) Assessment of the cPAS-based BGISEQ-500 platform for metagenomic sequencing. Gigascience 7:1–8
Chang L, Yu H, Miao X, Zhang J, Li S (2019) Development and comprehensive evaluation of a noninvasive prenatal paternity testing method through a scaled trial. Forensic Sci Int Genet 43:102158
SWGDAM (2012) SWGDAM validation guidelines for DNA analysis methods. 1-15. Available at https://docs.wixstatic.com/ugd/4344b0_813b241e8944497e99b9c45b163b76bd.pdf. Accessed 10 May 2020
Woerner AE, King JL, Budowle B (2017) Fast STR allele identification with STRait Razor 3.0. Forensic Sci Int Genet 30:18–23
Li R, Wu R, Li H, Zhang Y, Peng D, Wang N, Shen X, Wang Z, Sun H (2020) Characterizing stutter variants in forensic STRs with massively parallel sequencing. Forensic Sci Int Genet 45:102225
Bieber FR, Buckleton JS, Budowle B, Butler JM, Coble MD (2016) Evaluation of forensic DNA mixture evidence: protocol for evaluation, interpretation, and statistical calculations using the combined probability of inclusion. BMC Genet 17:125
Gill P (2001) An assessment of the utility of single nucleotide polymorphisms (SNPs) for forensic purposes. Int J Legal Med 114:204–210
Li R, Li H, Peng D, Hao B, Wang Z, Huang E, Wu R, Sun H (2019) Improved pairwise kinship analysis using massively parallel sequencing. Forensic Sci Int Genet 38:77–85
Davis C, Peters D, Warshauer D et al (2015) Sequencing the hypervariable regions of human mitochondrial DNA using massively parallel sequencing: Enhanced data acquisition for DNA samples encountered in forensic testing. Leg Med 17:123–127
Kim H, Erlich HA, Calloway CD (2015) Analysis of mixtures using next generation sequencing of mitochondrial DNA hypervariable regions. Croat Med J 56:208–217
Lutz S, Weisser HJ, Heizmann J, Pollak S (1998) Location and frequency of polymorphic positions in the mtDNA control region of individuals from Germany. Int J Legal Med 111:67–77
Prieto L, Zimmermann B, Goios A, Rodriguez-Monge A, Paneto GG, Alves C, Alonso A, Fridman C, Cardoso S, Lima G, Anjos MJ, Whittle MR, Montesino M, Cicarelli RMB, Rocha AM, Albarrán C, de Pancorbo MM, Pinheiro MF, Carvalho M, Sumita DR, Parson W (2011) The GHEP-EMPOP collaboration on mtDNA population data - a new resource for forensic casework. Forensic Sci Int Genet 5:146–151
Parson W, Gusmão L, Hares DR, Irwin JA, Mayr WR, Morling N, Pokorak E, Prinz M, Salas A, Schneider PM, Parsons TJ, DNA Commission of the International Society for Forensic Genetics (2014) DNA Commission of the International Society for Forensic Genetics: revised and extended guidelines for mitochondrial DNA typing. Forensic Sci Int Genet 13:134–142
Bandelt HJ, Van Oven M, Salas A (2012) Haplogrouping mitochondrial DNA sequences in legal medicine/forensic genetics. Int J Legal Med 126:901–916
Ensenberger MG, Lenz KA, Matthies LK, Hadinoto GM, Schienman JE, Przech AJ, Morganti MW, Renstrom DT, Baker VM, Gawrys KM, Hoogendoorn M, Steffen CR, Martín P, Alonso A, Olson HR, Sprecher CJ, Storts DR (2016) Developmental validation of the PowerPlex® fusion 6C system. Forensic Sci Int Genet 21:134–144
Wang DY, Gopinath S, Lagacé RE, Norona W, Hennessy LK, Short ML, Mulero JJ (2015) Developmental validation of the GlobalFiler® Express PCR Amplification Kit: a 6-dye multiplex assay for the direct amplification of reference samples. Forensic Sci Int Genet 19:148–155
Wang Z, Zhou D, Jia Z, Li L, Wu W, Li C, Hou Y (2016) Developmental validation of the Huaxia Platinum System and application in 3 main ethnic groups of China. Sci Rep 6:31075
Guo F, Zhou Y, Song H, Zhao J, Shen H, Zhao B, Liu F, Jiang X (2016) Next generation sequencing of SNPs using the HID-Ion AmpliSeq™ Identity Panel on the Ion Torrent PGM™ platform. Forensic Sci Int Genet 25:73–84
Novroski NMM, King JL, Churchill JD, Seah LH, Budowle B (2016) Characterization of genetic sequence variation of 58 STR loci in four major population groups. Forensic Sci Int Genet 25:214–226
Churchill JD, Novroski NMM, King JL, Seah LH, Budowle B (2017) Population and performance analyses of four major populations with Illumina’s FGx Forensic Genomics System. Forensic Sci Int Genet 30:81–92
Acknowledgments
The authors would like to thank Yicong Wang, Ning Wang, Wen Hu, Qiang Hou, and Fang Chen from MGI, BGI-Shenzhen for their technical assistance in support of this work. We would also like to thank LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (81971798, 81671873) and the Natural Science Foundation of Guangdong Province (2019A15150).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Informed consents were obtained before sample collection. For ethical approval, please refer to Supplementary Material Ethical Approval.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, R., Shen, X., Chen, H. et al. Developmental validation of the MGIEasy Signature Identification Library Prep Kit, an all-in-one multiplex system for forensic applications. Int J Legal Med 135, 739–753 (2021). https://doi.org/10.1007/s00414-021-02507-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-021-02507-0