Abstract
Main conclusion
DNA methylation of anthocyanin biosynthesis-related genes and MYB/bHLH transcription factors was associated with apple fruit skin color revealed by whole-genome bisulfite sequencing.
Abstract
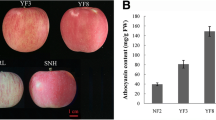
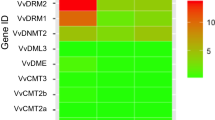
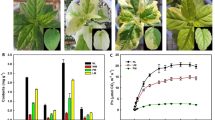
DNA methylation is a common feature of epigenetic regulation and is associated with various biological processes. Anthocyanins are among the secondary metabolites that contribute to fruit colour, which is a key appearance and nutrition quality attribute of apple fruit. Although few studies reported that DNA methylation in the promoter of MYB transcription factor was associated with fruit skin color, there is a general lack of understanding of the dynamics of global and genic DNA methylation in apple fruit. Here, whole-genome bisulfite sequencing was carried out in fruit skin of apple (Malus domestica Borkh.) cv. ‘Red Delicious’ (G0) and its four-generation bud sport mutants, including ‘Starking Red’ (G1), ‘Starkrimson’ (G2), ‘Campbell Redchief’ (G3) and ‘Vallee spur’ (G4) at color break stage. Correlation and linear-regression analysis between DNA methylation level and anthocyanin content, as well as the transcription levels of genes related to anthocyanin biosynthesis were carried out. The results showed that the number of differentially methylated regions (DMRs) and differentially methylated genes (DMGs) was considerably increased from G1 to G4 versus the number observed in G0. The mCHH context was dominant in apple, but the levels of mCG and mCHG of DMGs were significantly higher than that of the mCHH. Genetic variation of bud sport mutants from ‘Red Delicious’ was associated with differential DNA methylation. Additionally, hypomethylation of mCG and mCHG contexts in flavonoid biosynthesis pathway genes (PAL, 4CL, CYP98A, PER, CCoAOMT, CHS, and F3′H), mCHG context in MYB10 at upstream, led to transcriptional activation and was conductive to anthocyanin accumulation. However, hypermethylation of mCG context in bHLH74 at upstream led to transcriptional inhibition, inhibiting anthocyanin accumulation.








Similar content being viewed by others
Abbreviations
- DMGs:
-
Different methylation genes
- DMRs:
-
Different methylation regions
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Arrizabalaga M, Morales F, Oyarzun M, Delrot S, Gomès E, Irigoyen JJ, Hilbert G, Pascual I (2018) Tempranillo clones differ in the response of berry sugar and anthocyanin accumulation to elevated temperature. Plant Sci 267:74–83
Boyer J, Liu RH (2004) Apple phytochemicals and their health benefits. Nutr J 3:5
Candaele J, Demuynck K, Mosoti D, Beemster GTS, Inze D, Nelissen H (2014) Differential methylation during maize leaf growth targets developmentally regulated genes. Plant Physiol 164(3):1350–1364
Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99(Suppl. 4):16491–16498
Cedar H, Bergman Y (2009) Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet 10:295–304
Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6:351–360
Chopra S, Cocciolone SM, Bushman S, Sangar V, McMullen MD, Peterson T (2003) The maize Unstable factor for orange1 is a dominant epigenetic modifier of a tissue specifically silent allele of pericarp color1. Genetics 163:1135–1146
Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C (2008) Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 165(8):886–894
Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R et al (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49(7):1099–1106
D’Amato F (1997) Role of somatic mutations in the evolution of higher plants. Caryologia 50(1):1–15
El-Sharkawy I, Liang D, Xu K (2015) Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J Exp Bot 66(22):7359–7376
Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93:8449–8454
Gill DE, Chao L, Perkins SL, Wolf JB (1995) Genetic mosaicism in plants and clonal animals. Annu Rev Ecol S 26:423–444
Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74(1):481–514
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Honda C, Moriya S (2018) Anthocyanin biosynthesis in apple fruit. Horticult J 87:305–314
Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa AK, Zhu JK (2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci USA 114:E4511–E4519
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11(3):204–220
Lei M, Zhang H, Julian R, Tang K, Xie S, Zhu JK (2015) Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA 112(11):3553
Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM, Lisch D, Schmitz RJ, Vaughn MW, Springer NM (2015) RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci USA 112:14728–14733
Li Y, Dong X, Wang X, He T, Zhang H, Yang L, Wang T, Chen L, Gai J, Yang S (2017) Genome-wide comparative analysis of DNA methylation between soybean cytoplasmic male-sterile line NJCMS5A and its maintainer NJCMS5B. BMC Genomics 18:596
Li WF, Mao J, Yang SJ, Guo ZG, Ma ZH, Dawuda MM, Zuo CW, Chu MY, Chen BH (2018) Anthocyanin accumulation correlates with hormones in the fruit skin of ‘Red Delicious’ and its four generation bud sport mutants. BMC Plant Biol 18(1):363
Massonnet M, Fasoli M, Tornielli GB, Altieri M, Sandri M, Zuccolotto P, Paci P, Gardiman M, Zenoni S, Pezzott M (2017) Ripening transcriptomic program in red and white grapevine varieties correlates with berry skin anthocyanin accumulation. Plant Physiol 174(4):2376–2396
Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130(5):851–862
Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet 15(6):394–408
Pecinka A, Abdelsamad A, Vu GT (2013) Hidden genetic nature of epigenetic natural variation in plants. Trends Plant Sci 18(11):625–632
Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree. Annu Rev Ecol Evol S 37:187–214
Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181:219–229
Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen, H, Schork NJ, Schork JR (2013) Patterns of population epigenomic diversity. Nature 495(7440):193–198
Schob H, Grossniklaus U (2006) The first high-resolution DNA ‘‘methylome’’. Cell 126:1025–1028
Schultz MD, Schmitz RJ, Ecker JR (2012) ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet 28:583–585
Sekhon RS, Chopra S (2008) Progressive loss of DNA methylation releases epigenetic gene silencing from a tandemly repeated maize Myb gene. Genetics 181(1):81–91
Song Y, Ma K, Ci D, Chen Q, Tian J, Zhang D (2013) Sexual dimorphic floral development in dioecious plants revealed by transcriptome, phytohormone, and DNA methylation analysis in Populus tomentosa. Plant Mol Biol 83:559–576
Sun D, Xi Y, Rodriguez B, Park HJ, Tong P, Meong M, Goodell MA, Li W (2014) MOABS: model based analysis of bisulfite sequencing data. Genome Biol 15(2):R38
Telias A, Bradeen JM, Luby JJ, Hoover EE, Allan AC (2011a) Regulation of anthocyanin accumulation in apple peel. Hortic Rev 38:357–391
Telias A, Lin-Wang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM (2011b) Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol 11(1):93
Torregrosa L, Fernandez L, Bouquet A, Boursiquot JM, Pelsy F, Martinez-Zapater JM (2011) Origins and consequences of somatic variation in grapevine. Science Publishers, New Hampshire, pp 68–92
Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013) The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol 162(2):885–896
Williams RS, Benkeblia N (2018) Biochemical and physiological changes of star apple fruit (Chrysophyllum cainito), during different “on plant” maturation and ripening stages. Sci Hortic 236:36–42
Wilson GG, Murray NE (1991) Restriction and modification systems. Annu Rev Genet 25(1):585–627
Xu Y, Feng S, Jiao Q, Liu C, Zhang W, Chen W, Chen X (2012) Comparison of MdMYB1 sequences and expression of anthocyanin biosynthetic and regulatory genes between Malus domestica Borkh. cultivar ‘Ralls’ and its blushed sport. Euphytica 185(2):157–170
Yaish MW, Al-Lawati A, Al-Harrasi I, Patankar HV (2018) Genome-wide DNA methylation analysis in response to salinity in the model plant caliph medic (Medicago truncatula). BMC Genomics 19(1):78
Yang H, Chang F, You C, Cui J, Zhu G, Wang L, Zheng Y, Qi J, Ma H (2015) Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis. Plant J 81(2):268–281
Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919
Zhang H, Zhu J-K (2012a) Active DNA demethylation in plants and animals. Cold Spring Harb Symp Quant Biol 77:161–173
Zhang H, Zhu J-K (2012b) Seeing the forest for the trees: a wide perspective on RNA-directed DNA methylation. Genes Dev 26(16):1769–1773
Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126(6):1189–1201
Zhang Q, Wang D, Lang Z, He L, Yang L, Zeng L, Li Y, Zhao C, Huang H, Zhang H, Zhang H, Zhu J-K (2016) Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci USA 113(29):201607851
Zhang H, Lang Z, Zhu J-K (2018) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19(8):489–506
Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39:61–69
Acknowledgements
This work was financially supported by the Discipline Construction Fund Project of Gansu Agricultural University (GSAU-XKJS-2018-230), the Fostering Foundation for the Excellent Ph.D. Dissertation of Gansu Agricultural University (2017002), and the Science and Technology Major Project of Gansu Province (18ZD2NA006).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
DNA methylation levels of annotated DMGs involved in phenylpropanoid/flavonoid biosynthesis pathway in regions of a upstream, b genebody and c downstream used as a test group in total methylation level
Rights and permissions
About this article
Cite this article
Li, WF., Ning, GX., Mao, J. et al. Whole-genome DNA methylation patterns and complex associations with gene expression associated with anthocyanin biosynthesis in apple fruit skin. Planta 250, 1833–1847 (2019). https://doi.org/10.1007/s00425-019-03266-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-019-03266-4