Abstract
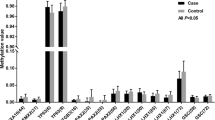
An imprinting disorder has been believed to underlie the etiology of familial biparental hydatidiform moles (HMs) based on the abnormal methylation or expression of imprinted genes in molar tissues. However, the extent of the epigenetic defect in these tissues and the developmental stage at which the disorder begins have been poorly defined. In this study, we assessed the extent of abnormal DNA methylation in two HMs caused by mutations in the recently identified 19q13.4 gene, NALP7. We demonstrate normal postzygotic DNA methylation patterns at major repetitive and long interspersed nuclear elements (LINEs), genes on the inactive X-chromosome, three-cancer related genes, and CpG rich regions surrounding the PEG3 differentially methylated region (DMR). Our data provide a comprehensive assessment of DNA methylation in familial molar tissues and indicate that abnormal DNA methylation in these tissues is restricted to imprinted DMRs. The known role of NALP7 in apoptosis and inflammation pinpoints previously unrecognized pathways that could directly or indirectly underlie the abnormal methylation of imprinted genes in molar tissues.




Similar content being viewed by others
References
Agulnik AI, Mitchell MJ, Mattei MG, Borsani G, Avner PA, Lerner JL, Bishop CE (1994) A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum Mol Genet 3:879–884
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51:1229–1239
Asakawa J, Kuick R, Neel JV, Kodaira M, Satoh C, Hanash SM (1995) Quantitative and qualitative genetic variation in two-dimensional DNA gels of human lymphocytoid cell lines. Electrophoresis 16:241–252
Beever C, Lai BP, Baldry SE, Penaherrera MS, Jiang R, Robinson WP, Brown CJ (2003) Methylation of ZNF261 as an assay for determining X chromosome inactivation patterns. Am J Med Genet A 120:439–441
Bird AP (1986) CpG-rich islands and the function of DNA methylation. Nature 321:209–213
Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38–44
El-Maarri O, Herbiniaux U, Walter J, Oldenburg J (2002) A rapid, quantitative, non-radioactive bisulfite-SNuPE- IP RP HPLC assay for methylation analysis at specific CpG sites. Nucleic Acids Res 30:e25
El-Maarri O, Seoud M, Coullin P, Herbiniaux U, Oldenburg J, Rouleau G, Slim R (2003) Maternal alleles acquiring paternal methylation patterns in biparental complete hydatidiform moles. Hum Mol Genet 12:1405–1413
El-Maarri O, Seoud M, Riviere JB, Oldenburg J, Walter J, Rouleau G, Slim R (2005) Patients with familial biparental hydatidiform moles have normal methylation at imprinted genes. Eur J Hum Genet 13:486–490
Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM (2003) Predicting aberrant CpG island methylation. Proc Natl Acad Sci USA 100:12253–12258
Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE (2002) Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet 31:175–179
Gilbert SL, Sharp PA (1999) Promoter-specific hypoacetylation of X-inactivated genes. Proc Natl Acad Sci USA 96:13825–13830
Hansen RS (2003) X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Hum Mol Genet 12:2559–2567
Hatada I, Fukasawa M, Kimura M, Morita S, Yamada K, Yoshikawa T, Yamanaka S, Endo C, Sakurada A, Sato M, Kondo T, Horii A, Ushijima T, Sasaki H (2006) Genome-wide profiling of promoter methylation in human. Oncogene
Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R (2001) Epigenetic instability in ES cells and cloned mice. Science 293:95–97
Judson H, Hayward BE, Sheridan E, Bonthron DT (2002) A global disorder of imprinting in the human female germ line. Nature 416:539–542
Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, Macdonald F, Sampson JR, Barratt CL, Reik W, Hawkins MM (2003) Beckwith–Wiedemann syndrome and assisted reproduction technology (ART). J Med Genet 40:62–64
Mann MR, Bartolomei MS (2002) Epigenetic reprogramming in the mammalian embryo: struggle of the clones. Genome Biol 3:REVIEWS1003
McMaster MT, Dey SK, Andrews GK (1993) Association of monocytes and neutrophils with early events of blastocyst implantation in mice. J Reprod Fertil 99:561–569
Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, Hanash S, Rouleau GA, Slim R (2006) Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 38:300–302
Reik W, Walter J (2001) Genomic imprinting: parental influence on the genome. Nat Rev Genet 2:21–32
Rivier C, Vale W (1990) Cytokines act within the brain to inhibit luteinizing hormone secretion and ovulation in the rat. Endocrinology 127:849–856
Santos F, Hendrich B, Reik W, Dean W (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241:172–182
Smiraglia DJ, Plass C (2002) The study of aberrant methylation in cancer via restriction landmark genomic scanning. Oncogene 21:5414–5426
Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA (2005) Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci USA 102:3336–3341
Strakova Z, Mavrogianis P, Meng X, Hastings JM, Jackson KS, Cameo P, Brudney A, Knight O, Fazleabas AT (2005) In vivo infusion of interleukin-1beta and chorionic gonadotropin induces endometrial changes that mimic early pregnancy events in the baboon. Endocrinology 146:4097–4104
Vrana PB, Guan XJ, Ingram RS, Tilghman SM (1998) Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nat Genet 20:362–365
Wongweragiat S, Searle RF, Bulmer JN (1999) Decidual T lymphocyte activation in hydatidiform mole. J Clin Pathol 52:888–894
Wrenzycki C, Herrmann D, Lucas-Hahn A, Gebert C, Korsawe K, Lemme E, Carnwath JW, Niemann H (2005) Epigenetic reprogramming throughout preimplantation development and consequences for assisted reproductive technologies. Birth Defects Res C Embryo Today 75:1–9
Xue WC, Chan KY, Feng HC, Chiu PM, Ngan HY, Tsao SW, Cheung AN (2004) Promoter hypermethylation of multiple genes in hydatidiform mole and choriocarcinoma. J Mol Diagn 6:326–334
Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13:335–340
Acknowledgments
We would like to thank the members of the families for their participation in this study, Dr. Paul Fournier for his help in obtaining trophoblast tissues from therapeutic abortions. R.S. is supported by the Fonds de la Recherche en Santé du Québec and by an operating (MOP-67179) and an international development (OPD-73018) grants from the Canadian Institute of Health Research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Djuric, U., El-Maarri, O., Lamb, B. et al. Familial molar tissues due to mutations in the inflammatory gene, NALP7, have normal postzygotic DNA methylation. Hum Genet 120, 390–395 (2006). https://doi.org/10.1007/s00439-006-0192-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-006-0192-3