Abstract
This paper presents a new evolutionary cooperative–competitive algorithm for the design of Radial Basis Function Networks (RBFNs) for classification problems. The algorithm, CO2RBFN, promotes a cooperative–competitive environment where each individual represents a radial basis function (RBF) and the entire population is responsible for the final solution. The proposal considers, in order to measure the credit assignment of an individual, three factors: contribution to the output of the complete RBFN, local error and overlapping. In addition, to decide the operators’ application probability over an RBF, the algorithm uses a Fuzzy Rule Based System. It must be highlighted that the evolutionary algorithm considers a distance measure which deals, without loss of information, with differences between nominal features which are very usual in classification problems. The precision and complexity of the network obtained by the algorithm are compared with those obtained by different soft computing methods through statistical tests. This study shows that CO2RBFN obtains RBFNs with an appropriate balance between accuracy and simplicity, outperforming the other methods considered.




Similar content being viewed by others
References
Alcalá-Fdez J, Sánchez L, García S, Del Jesus MJ, Ventura S, Garrell JM, Otero J, Romero C, Bacardit J, Rivas VM, Fernández JC, Herrera F (2009) KEEL: a software tool to assess evolutionary algorithms to data mining problems. Soft Comput 13(3): 307–318
Ampazis N, Perantonis SJ (2002) Two highly efficient second-order algorithms for training feedforwards networks. IEEE Trans Neural Netw 13(3):1064–1074
Asuncion A, Newman DJ (2007) UCI machine learning repository. School of Information and Computer Science, University of California, Irvine, CA. http://www.ics.uci.edu/~mlearn/MLRepository.html
Bäck T, Hammel U, Schwefel H (1997) Evolutionary computation: comments on the history and current state. IEEE Trans Evol Comput 1(1):3–17
Balakrishnan K, Honavar V (1995) Evolutionary design of neural architecture—a preliminary taxonomy and guide to literature. Technical report, AI Research Group, CS-TR 95–01
Billings SA, Zheng GL (1995) Radial basis function network configuration using genetic algorithms. Neural Netw 8(6):877–890
Broomhead D, Lowe D (1988) Multivariable functional interpolation and adaptive networks. Complex Syst 2:321–355
Buchtala O, Klimek M, Sick B (2005) Evolutionary optimization of radial basis function classifiers for data mining applications. IEEE Trans Syst Man Cybern B 35(5):928–947
Burdsall B, Giraud-Carrier C (1997) GA-RBF: a self optimising RBF network. In: Proceedings of conference on artificial neural networks and genetic algorithms. Springer, Berlin
Chaiyaratana N, Zalzala AMS (1998) Evolving hybrid RBF-MLP networks using combined genetic/unsupervised/supervised learning. In: Proceedings of UKACC international conference on control, Swansea, UK
Chen S, Billings SA, Cowan CFN, Grant PW (1990) Practical identification of narmax models using radial basis functions. Int J Control 52(6):1327–1350
Chen S, Cowan C, Grant P (1991) Orthogonal least squares learning algorithm for radial basis function networks. IEEE Trans Neural Netw 2:302–309
Chen S, Wu Y, Luk BL (1999) Combined genetic algorithm optimization and regularized orthogonal least squares learning for radial basis function networks. IEEE Trans Neural Netw 10(5):1239–1243
Cheng V, Li CH, Kwok JT, Li CK (2004) Dissimilarity learning for nominal data. Pattern Recogn 37:1471–1477
Dawson CW, Wilby RL, Harpham C, Brown MR, Cranston, E, Darby EJ (2000) Modelling Ranunculus presence in the Rivers test and Itchen using artificial neural networks. In: Proceedings of international conference on geocomputation, Greenwich, UK
Deb K (2001) Multi-objective optimization using evolutionary algorithms, 1st edn. Wiley, New York
Demšar J (2006) Statistical Comparisons of Classifiers over Multiple Data Sets. J Mach Learn Res 7:1–30
Du H, Zhang N (2008) Time series prediction using evolving radial basis function networks with new enconding scheme. Neurocomputing 71:1388–1400
Er MJ, Chen W, Wu S (2005) High-speed face recognition base on discrete cosine transform and RBF neural networks. IEEE Trans Neural Netw 16(3):679–691
Esposito F, Malerba D, Tamma V, Bock HH (2000a) Classical resemblance measures. In: Bock H-H, Diday E (eds) Analysis of symbolic data. Exploratory methods for extracting statistical information from complex data, Series: studies in classification, data analysis, and knowledge organization, vol 15. Springer, Berlin, pp 139–152
Esposito A, Marinaro M, Oricchio D, Scarpetta S (2000b) Approximation of continuous and discontinuous mappings by a growing neural RBF-based algorithm. Neural Netw 13(6):651–665
Ferreira PM, Ruano AE, Fonseca CM (2003) Genetic assisted selection of RBF model structures for greenhouse inside air temperature prediction. IEEE Control Appl 1:576–581
Fogel LJ, Owens AJ, Walsh MJ (1966) Artificial Intelligence through simulation evolution. Wiley, New York
Fu X, Wang L (2002) A GA-based novel RBF classifier with class-dependent features. Proc Congr Evol Comput 2:1964–1969
Fu X, Wang L (2003) Data dimensionality reduction with application to simplifying RBF network structure and improving classification performance. IEEE Trans Syst Man Cybern B 33(3):399–409
Goldberg D (1989) Genetic algorithms in search, optimization, and machine learning. Addison-Wesley, Reading
Goldberg D, Richardson J (1987) Genetic algorithms with sharing for multimodal function optimization. In: Grefenstette (ed) Proceedings of second international conference on genetic algorithms. Lawrence Erlbaum Associates, pp 41–49
Golub G, Van Loan C (1996) Matrix computations, 3rd edn. J. Hopkins University Press, Baltimore
González J, Rojas I, Ortega J, Pomares H, Fernández FJ, Díaz AF (2003) Multiobjective evolutionary optimization of the size, shape, and position parameters of radial basis function networks for function approximation. IEEE Trans Neural Netw 14(6):1478–1495
Guillén A, Pomares H, Rojas I, González J, Herrera LJ, Rojas F, Valenzuela O (2007) Output value-based initialization for radial basis function neural networks. Neural Process Lett. doi:10.1007/s11063-007-9039-8
Harpham C, Dawson C, Brown M (2004) A review of genetic algorithms applied to training radial basis function networks. Neural Comput Appl 13:193–201
Holcomb T, Morari M (1991) Local training for radial basis function networks: towards solving the hidden unit problem. In: Proceedings of American control conference, Boston
Holland JH (1975) Adaptation in natural and artificial systems. The University of Michigan Press
Huang SN, Tan KK, Lee TH (2008) Adaptive neural network algorithm for control design of rigid-link electrically driven robots. Neurocomputing 71(4–6):885–894
Jang JSR, Sun CT (1993) Functional equivalence between radial basis functions and fuzzy inference systems. IEEE Trans Neural Netw 4:156–158
Jiang N, Zhao ZY, Ren LQ (2003) Design of structural modular neural networks with genetic algorithm. Adv Eng Soft 1:17–24
Jin Y, Sendhoff B (2003) Extracting interpretable fuzzy rules from RBF networks. Neural Process Lett 17(2):149–164
Lacerda E, Carvalho A, Braga A, Ludermir T (2005) Evolutionary radial functions for credit assessment. Appl Intell 22:167–181
Lee S, Kil RM (1991) A Gaussian potential function network with hierarchically seft-organising learning. Neural Netw 4:207–224
Leung H, Dubash N, Xie N (2002) Detection of small objects in clutter using a GA-RBF neural network. IEEE Trans Aero Electr Sys 38(1):98–118
Li M, Tian J, Chen F (2008) Improving multiclass pattern recognition with a co-evolutionary RBFNN. Pattern Recogn Lett 29(4):392–406
Maglogiannis I, Sarimveis H, Kiranoudis CT, Chatziioannou AA, Oikonomou N, Aidinis V (2008) Radial basis function neural networks classification for the recognition of idiopathic pulmonary fibrosis in microscopic images. IEEE Trans Inf Technol B 12(1):42–54
Mandani E, Assilian S (1975) An experiment in linguistic synthesis with a fuzzy logic controller. Int J Man Mach Stud 7(1):1–13
Marcos JV, Hornero R, Álvarez D, Del Campo F, López M, Zamarrón C (2008) Radial basis function classifiers to help in the diagnosis of the obstructive sleep apnoea syndrome from nocturnal oximetry. Med Biol Eng Comput 46:323–332
Moechtar M, Farag AS, Hu L, Cheng TC (1999) Combined genetic algorithms and neural network approach for power system transient stability evaluation. Europ Trans Elect Power 9(2):115–122
Moody J, Darken CJ (1989) Fast learning in networks of locally-tuned processing units. Neural Comput 1:281–294
Musavi MT, Ahmed W, Chan KH, Faris KB, Hummels DM (1992) On the training of radial basis function classifiers. Neural Netw 5:595–603
Neruda R, Kudová P (2005) Learning methods for radial basis function networks. Future Gener Comp Sy 21(7):1131–1142
Orr MJL (1995) Regularization on the selection of radial basis function centers. Neural Comput 7:606–623
Park J, Sandberg I (1991) Universal approximation using radial-basis function networks. Neural Comput 3:246–257
Park J, Sandberg I (1993) Universal approximation and radial basis function network. Neural Comput 5(2):305–316
Pedrycz W (1998) Conditional fuzzy clustering in the design of radial basis function neural networks. IEEE Trans Neural Netw 9(4):601–612
Peng JX, Li K, Huang DS (2006) A hybrid forward algorithm for RBF neural network construction. IEEE Trans Neural Netw 17(6):1439–1451
Pérez-Godoy MD, Rivera AJ, del Jesus MJ, Rojas I (2007) CoEvRBFN: an approach to solving the classification problem with a hybrid cooperative–coevolutive algorithm. In: Proceedings of international workshop on artificial neural networks, pp 324–332
Pérez-Godoy MD, Aguilera JJ, Berlanga FJ, Rivas VM, Rivera AJ (2008) A preliminary study of the effect of feature selection in evolutionary RBFN design. In: Proceedings of information processing and management of uncertainty in knowledge-based system, pp 1151–1158
Plat J (1991) A resource allocating network for function interpolation. Neural Comput 3(2):213–225
Potter M, De Jong K (2000) Cooperative coevolution: an architecture for evolving coadapted subcomponents. Evol Comput 8(1):1–29
Powell M (1985) Radial basis functions for multivariable interpolation: a review. In: IMA Proceedings of conference on algorithms for the approximation of functions and data, pp 143–167
Quinlan JR (1993) C4.5: programs for machine learning. Morgan Kauffman, Menlo Park
Rivas VM, Merelo JJ, Castillo PA, Arenas MG, Castellanos JG (2004) Evolving RBF neural networks for time-series forecasting with EvRBF. Inf Sci 165(3–4):207–220
Rivera AJ, Ortega J, Prieto A (2001) Design of RBF networks by cooperative/competitive evolution of units. In: Proceedings of international conference on artificial neural networks and genetic algorithms (ICANNGA 2001), pp 375–378
Rivera AJ, Rojas I, Ortega J, del Jesus MJ (2007) A new hybrid methodology for cooperative–coevolutionary optimization of radial basis function networks. Soft Comput. doi:10.1007/s00500-006-0128-9
Rojas R, Feldman J (1996) Neural networks: a systematic introduction. Springer, Berlin
Rojas I, Valenzuela O, Prieto A (1997) Statisctical analysis of the main parameters in the definition of radial basis function networks. Lect Notes Comput Sci 1240:882–891
Sanchez VD (2002) A searching for a solution to the automatic RBF network design problem. Neurocomputing 42:147–170
Schaffer JD, Whitley D, Eschleman LJ (1992) Combinations of genetic algorithms and neural networks: a survey of the state of the art. In: Proceedings of international workshop on combinations of genetic algorithms and neural networks
Sergeev SA, Mahotilo KV, Voronovsky GK, Petrashev SN (1998) Genetic algorithm for training dynamical object emulator based on RBF neural network. Int J Appl Electro Mech 9(1):65–74
Sheta AF, De Jong K (2001) Time-series forecasting using GA-tuned radial basis functions. Info Sci 133(3–4):221–228
Stanfill C, Waltz D (1986) Towards memory-based reasoning, Commun. ACM 29(12):1213–1228
Sumathi S, Sivanandam SN, Ravindran R (2001) Design of a soft computing hybrid model classifier for data mining applications. Engin Intell Sys Electr Engin Comm 9(1):33–56
Sun YF, Liang YC, Zhang WL, Lee HP, Lin WZ, Cao LJ (2005) Optimal partition algorithm of the RBF neural network and its application to financial time series forecasting. Neural Comput Appl 14(1):36–44
Sundararajan N, Saratchandran P, Yingwei L (1999) Radial basis function neural network with sequential learning: MRAN and its application. World Scientifics, New York
Teixeira CA, Ruano MG, Ruano AE, Pereira WCA (2008) A soft-computing methodology for noninvasive time-spatial temperature estimation. IEEE Trans Bio Med Eng 55(2):572–580
Topchy A, Lebedko O, Miagkikh V, Kasabov N (1997) Adaptive training of radial basis function networks based on co-operative evolution and evolutionary programming. In: Proceedings of international conference neural information processing (ICONIP), pp 253–258
Vesin JM, Gruter R (1999) Model selection using a simplex reproduction genetic algorithm. Sig Process 78:321–327
Whitehead B, Choate T (1996) Cooperative–competitive genetic evolution of radial basis function centers and widths for time series prediction. IEEE Trans Neural Netw 7(4):869–880
Widrow B, Lehr MA (1990) 30 Years of adaptive neural networks: perceptron, madaline and backpropagation. Proc IEEE 78(9):1415–1442
Wilson DR, Martinez TR (1997) Improved heterogeneous distance functions. J Artif Intell Res 6(1):1–34
Xue Y, Watton J (1998) Dynamics modelling of fluid power systems applying a global error descent algorithm to a selforganising radial basis function network. Mechatronics 8(7):727–745
Yao X (1993) A review of evolutionary artificial neural networks. Int J Intell Syst 8(4):539–567
Yao X (1999) Evolving artificial neural networks. Proc IEEE 87(9):1423–1447
Yen GG (2006) Multi-Objective evolutionary algorithm for radial basis function neural network design. Stud Comput Intell 16:221–239
Acknowledgments
This work has been partially supported by the CICYT Spanish Projects TIN2005-04386-C05-03, TIN2007-60587 and the Andalusian Research Plan TIC-3928.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: Detailed results obtained for CO2RBFN
Car dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
4 | 0.129 | 0.007 | 0.207 | 0.061 | 87.104 | 79.350 |
5 | 0.122 | 0.007 | 0.190 | 0.048 | 87.823 | 81.007 |
6 | 0.116 | 0.008 | 0.204 | 0.045 | 88.354 | 79.572 |
7 | 0.112 | 0.007 | 0.198 | 0.046 | 88.753 | 80.197 |
8 | 0.108 | 0.009 | 0.198 | 0.051 | 89.186 | 80.241 |
9 | 0.102 | 0.009 | 0.200 | 0.056 | 89.784 | 79.998 |
10 | 0.098 | 0.009 | 0.207 | 0.053 | 90.203 | 79.270 |
11 | 0.095 | 0.009 | 0.191 | 0.052 | 90.543 | 80.925 |
12 | 0.093 | 0.011 | 0.200 | 0.040 | 90.678 | 80.011 |
13 | 0.089 | 0.008 | 0.202 | 0.047 | 91.075 | 79.804 |
14 | 0.087 | 0.010 | 0.207 | 0.049 | 91.279 | 79.261 |
15 | 0.079 | 0.008 | 0.220 | 0.061 | 92.085 | 77.974 |
16 | 0.079 | 0.010 | 0.199 | 0.045 | 92.103 | 80.127 |
Credit dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
2 | 0.123 | 0.006 | 0.158 | 0.065 | 87.655 | 84.232 |
3 | 0.120 | 0.004 | 0.158 | 0.081 | 87.974 | 84.203 |
4 | 0.118 | 0.004 | 0.177 | 0.100 | 88.235 | 82.261 |
5 | 0.116 | 0.005 | 0.175 | 0.096 | 88.432 | 82.522 |
6 | 0.115 | 0.003 | 0.172 | 0.088 | 88.470 | 82.812 |
7 | 0.114 | 0.004 | 0.167 | 0.084 | 88.577 | 83.275 |
8 | 0.113 | 0.004 | 0.179 | 0.094 | 88.686 | 82.116 |
Glass dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
7 | 0.328 | 0.020 | 0.358 | 0.113 | 67.223 | 64.216 |
8 | 0.319 | 0.016 | 0.373 | 0.103 | 68.145 | 62.699 |
9 | 0.310 | 0.017 | 0.354 | 0.104 | 68.987 | 64.575 |
10 | 0.296 | 0.014 | 0.360 | 0.120 | 70.410 | 63.990 |
11 | 0.290 | 0.015 | 0.354 | 0.111 | 71.034 | 64.635 |
12 | 0.282 | 0.017 | 0.333 | 0.105 | 71.812 | 66.694 |
13 | 0.277 | 0.014 | 0.332 | 0.111 | 72.299 | 66.778 |
14 | 0.275 | 0.016 | 0.356 | 0.116 | 72.547 | 64.389 |
15 | 0.266 | 0.015 | 0.330 | 0.109 | 73.399 | 66.976 |
16 | 0.262 | 0.015 | 0.343 | 0.107 | 73.826 | 65.654 |
17 | 0.258 | 0.015 | 0.346 | 0.104 | 74.230 | 65.425 |
18 | 0.255 | 0.016 | 0.335 | 0.103 | 74.490 | 66.487 |
19 | 0.251 | 0.014 | 0.340 | 0.117 | 74.925 | 65.980 |
20 | 0.250 | 0.017 | 0.349 | 0.109 | 74.977 | 65.086 |
21 | 0.248 | 0.014 | 0.354 | 0.118 | 75.164 | 64.602 |
22 | 0.248 | 0.014 | 0.323 | 0.111 | 75.175 | 67.710 |
23 | 0.242 | 0.016 | 0.328 | 0.114 | 75.798 | 67.223 |
24 | 0.240 | 0.015 | 0.332 | 0.118 | 75.997 | 66.763 |
25 | 0.236 | 0.014 | 0.342 | 0.116 | 76.389 | 65.820 |
26 | 0.233 | 0.015 | 0.326 | 0.102 | 76.700 | 67.391 |
27 | 0.234 | 0.019 | 0.329 | 0.115 | 76.587 | 67.065 |
28 | 0.235 | 0.015 | 0.337 | 0.107 | 76.536 | 66.332 |
Hepatitis dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
2 | 0.087 | 0.016 | 0.168 | 0.145 | 91.270 | 83.228 |
3 | 0.074 | 0.009 | 0.168 | 0.138 | 92.632 | 83.157 |
4 | 0.067 | 0.011 | 0.151 | 0.094 | 93.261 | 84.905 |
5 | 0.065 | 0.011 | 0.151 | 0.071 | 93.548 | 84.914 |
6 | 0.064 | 0.008 | 0.128 | 0.107 | 93.563 | 87.187 |
7 | 0.063 | 0.010 | 0.139 | 0.075 | 93.749 | 86.137 |
8 | 0.057 | 0.007 | 0.126 | 0.074 | 94.280 | 87.399 |
Ionosphere dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
2 | 0.149 | 0.017 | 0.171 | 0.053 | 85.078 | 82.926 |
3 | 0.134 | 0.020 | 0.160 | 0.049 | 86.648 | 84.003 |
4 | 0.105 | 0.015 | 0.134 | 0.050 | 89.485 | 86.579 |
5 | 0.097 | 0.017 | 0.119 | 0.049 | 90.294 | 88.069 |
6 | 0.087 | 0.014 | 0.111 | 0.043 | 91.270 | 88.907 |
7 | 0.074 | 0.013 | 0.099 | 0.048 | 92.643 | 90.111 |
8 | 0.065 | 0.011 | 0.086 | 0.039 | 93.485 | 91.411 |
Iris dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
3 | 0.017 | 0.008 | 0.045 | 0.047 | 98.252 | 95.467 |
4 | 0.012 | 0.004 | 0.048 | 0.055 | 98.770 | 95.200 |
5 | 0.011 | 0.004 | 0.045 | 0.052 | 98.919 | 95.467 |
6 | 0.010 | 0.004 | 0.037 | 0.042 | 99.007 | 96.267 |
7 | 0.010 | 0.005 | 0.037 | 0.050 | 99.007 | 96.267 |
8 | 0.009 | 0.004 | 0.044 | 0.054 | 99.067 | 95.600 |
9 | 0.009 | 0.004 | 0.052 | 0.050 | 99.081 | 94.800 |
10 | 0.009 | 0.004 | 0.044 | 0.049 | 99.141 | 95.600 |
11 | 0.009 | 0.004 | 0.048 | 0.050 | 99.111 | 95.200 |
12 | 0.008 | 0.004 | 0.040 | 0.046 | 99.230 | 96.000 |
Pima dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
2 | 0.239 | 0.024 | 0.260 | 0.051 | 76.068 | 74.001 |
3 | 0.227 | 0.010 | 0.248 | 0.043 | 77.341 | 75.218 |
4 | 0.221 | 0.007 | 0.240 | 0.049 | 77.873 | 75.950 |
5 | 0.218 | 0.006 | 0.247 | 0.047 | 78.224 | 75.252 |
6 | 0.215 | 0.006 | 0.243 | 0.047 | 78.516 | 75.716 |
7 | 0.213 | 0.006 | 0.244 | 0.045 | 78.675 | 75.638 |
8 | 0.212 | 0.006 | 0.242 | 0.044 | 78.814 | 75.796 |
Sonar dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
2 | 0.247 | 0.020 | 0.282 | 0.111 | 75.299 | 71.762 |
3 | 0.235 | 0.019 | 0.261 | 0.114 | 76.517 | 73.910 |
4 | 0.222 | 0.015 | 0.283 | 0.097 | 77.756 | 71.705 |
5 | 0.219 | 0.020 | 0.285 | 0.092 | 78.120 | 71.514 |
6 | 0.210 | 0.013 | 0.279 | 0.099 | 78.952 | 72.114 |
7 | 0.206 | 0.016 | 0.271 | 0.090 | 79.412 | 72.948 |
8 | 0.201 | 0.012 | 0.249 | 0.098 | 79.882 | 75.086 |
Vehicle dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
4 | 0.432 | 0.021 | 0.446 | 0.048 | 56.782 | 55.391 |
5 | 0.400 | 0.019 | 0.415 | 0.054 | 60.032 | 58.489 |
6 | 0.389 | 0.021 | 0.405 | 0.054 | 61.135 | 59.520 |
7 | 0.369 | 0.026 | 0.381 | 0.045 | 63.092 | 61.912 |
8 | 0.357 | 0.019 | 0.386 | 0.043 | 64.303 | 61.390 |
9 | 0.343 | 0.016 | 0.370 | 0.050 | 65.721 | 62.954 |
10 | 0.332 | 0.016 | 0.350 | 0.047 | 66.840 | 64.979 |
11 | 0.318 | 0.015 | 0.353 | 0.046 | 68.201 | 64.703 |
12 | 0.311 | 0.013 | 0.334 | 0.045 | 68.873 | 66.645 |
13 | 0.307 | 0.010 | 0.326 | 0.041 | 69.320 | 67.414 |
14 | 0.296 | 0.014 | 0.318 | 0.038 | 70.433 | 68.201 |
15 | 0.292 | 0.014 | 0.316 | 0.044 | 70.846 | 68.433 |
16 | 0.287 | 0.012 | 0.308 | 0.047 | 71.253 | 69.192 |
Wbcd dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
2 | 0.024 | 0.004 | 0.032 | 0.023 | 97.581 | 96.769 |
3 | 0.023 | 0.002 | 0.033 | 0.021 | 97.743 | 96.741 |
4 | 0.023 | 0.002 | 0.033 | 0.021 | 97.749 | 96.739 |
5 | 0.022 | 0.002 | 0.029 | 0.018 | 97.797 | 97.083 |
6 | 0.022 | 0.002 | 0.032 | 0.019 | 97.794 | 96.792 |
7 | 0.022 | 0.002 | 0.033 | 0.020 | 97.803 | 96.740 |
8 | 0.022 | 0.002 | 0.029 | 0.020 | 97.813 | 97.054 |
Wine dataset
# Nodes | Error training | Standard dev | Error test | Stand dev | % Training | % Test |
|---|---|---|---|---|---|---|
3 | 0.008 | 0.006 | 0.051 | 0.060 | 99.189 | 94.915 |
4 | 0.004 | 0.004 | 0.057 | 0.066 | 99.575 | 94.261 |
5 | 0.003 | 0.003 | 0.043 | 0.061 | 99.738 | 95.732 |
6 | 0.002 | 0.003 | 0.038 | 0.045 | 99.813 | 96.157 |
7 | 0.001 | 0.002 | 0.033 | 0.046 | 99.913 | 96.739 |
8 | 0.001 | 0.002 | 0.036 | 0.040 | 99.950 | 96.412 |
9 | 0.000 | 0.001 | 0.037 | 0.046 | 99.975 | 96.281 |
10 | 0.000 | 0.000 | 0.045 | 0.054 | 100.000 | 95.484 |
11 | 0.000 | 0.001 | 0.038 | 0.047 | 99.975 | 96.196 |
12 | 0.000 | 0.000 | 0.038 | 0.050 | 100.000 | 96.190 |
Appendix 2: GeneticRBFN description
To test our cooperative–competitive method against a Pittsburgh based proposal, a typical genetic algorithm for the RBFN design, GeneticRBFN, has been developed. The design lines of GeneticRBFN are the classical ones for these kinds of algorithms (Harpham et al. 2004). In order to establish similar operating conditions certain characteristics of CO2RBFN have been introduced in GeneticRBFN, like analogies in the operators and the HVDM dissimilarity measure.
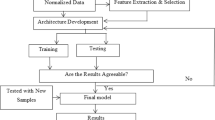
This method follows the traditional Pittsburgh evolutionary approach for the design of RBFNs: each individual is a whole network. The objective of the evolutionary process is to minimise the classification error. The main steps of this algorithm are shown in Fig. 5.
The main components of GeneticRBFN algorithm are described below.
2.1 Initialization
The initialization stage is the same as that in CO2RBFN. So the RBFs will be centred, in an equidistributed way, for each RBFN/individual.
2.2 Genetic operators: selection, recombination and mutation
With the crossover operator, two individuals/RBFNs parents are randomly chosen to obtain an RBFN offspring. The number of RBFs of the new individual will be delimited between a minimum and a maximum value. The minimum value is set to the number of RBFs of the parent with fewer RBFs. In the same way, the maximum value is set to the number of RBFs of the parent with more RBFs. In order to generate the offspring RBFs will be chosen in a random way from the parents.
Six mutation operators, usually considered in the specialised bibliography (Harpham et al. 2004) have been implemented. They can be classified as random operators or biased operators. The random operators are:
-
DelRandRBFs: randomly eliminates k RBFs, where k is a pm percent of the total number of RBFs in the RBFN.
-
InsRandRBFs: randomly aggregates k RBFs, where k is a pm percent of the total number of RBFs in the RBFN.
-
ModCentRBFs: randomly modifies the centre of k RBFs, where k is a pm percent of the total number of RBFs in the RBFN. The centre of the basis function will be modified in a pr percent of its width.
-
ModWidtRBFs: randomly modifies the centre of k RBFs, where k is a pm percent of the total number of RBFs in the RBFN. The width of the basis function will be modified in a pr percent of its width.
Biased operators, which exploit local information are:
-
DelInfRBFs: deletes the k RBFs of the RBFN with a lower weight. k is a pm percent of the total number of RBFs in the RBFN.
-
InsInfRBFs: inserts the k RBFs in the RBFN outside the width of any RBF present in the RBFN. k is a pm percent of the total number of RBFs in the RBFN.
An intermediate population with the parents and the offspring is considered and a tournament selection mechanism is used to determine the new population. The diversity of the population is promoted by using a low value for the tournament size (k = 3).
2.3 Training weights
In order to train the weights, the LMS algorithm is used. Its parameters are set to their standard values.
2.4 Individual evaluation
The fitness defined for each individual/RBFN is its classification error for the given problem.
In order to increase the efficiency of the GA, the search space of this method has been drastically reduced. As is well known, in Pittsburgh GAs, where the only objective to optimise is the classification error, the complexity of the individuals (i.e. number of RBFs) grows in an uncontrolled way (because normally an RBFN with more RBFs gives a lower error percentage than an RBFN with few RBFs). In this experimentation, the search space has been reduced by fixing the maximum complexity (and so chromosome size) between a minimum and a maximum number of RBFs. The minimum number of RBFs has been set to the number of classes for the problem and the maximum to four times this number.
Appendix 3: Statistical methods
Non-parametric methods are often referred to as distribution free methods, as they do not rely on assumptions that the data are drawn from a given probability distribution.
Non-parametric methods are widely used for studying populations which take a ranked order (such as movie reviews receiving one to four stars). The use of non-parametric methods may be necessary when data has a ranking but no clear numerical interpretation, such as when assessing preferences.
As non-parametric methods make fewer assumptions, their applicability is much wider than the corresponding parametric methods. In particular, they may be used in situations where less is known about the application in question. In addition, due to the reliance on fewer assumptions, non-parametric methods are more robust.
Another justification for the use of non-parametric methods is simplicity. In certain cases, even when the use of parametric methods is justified, non-parametric methods may be easier to use. Due both to this simplicity and to their greater robustness, non-parametric methods are seen by some statisticians as leaving less room for improper use and misunderstanding.
Now we will explain the non-parametric methods used:
3.1 Friedman’s test
This is a non-parametric equivalent of the test of repeated-measures ANOVA. It computes the ranking of the observed results for algorithm (r j for the algorithm j with k algorithms) for each data-set, assigning to the best the ranking 1, and to the worst the ranking k. Under the null hypothesis, formed from supposing that the results of the algorithms are equivalents and, therefore, their rankings are also similar, Friedman’s statistic
is distributed according to \( \chi_{F}^{2} \) with k − 1 degrees or freedom, being \( R_{j} = {\frac{1}{{N_{ds} }}}\sum\nolimits_{i} {r_{i}^{2} } , \) and N ds the number of data-set. The critical values for Friedman’s statistic coincide with those established in the \( \chi^{2} \) distribution when N ds > 10 and k > 5.
3.2 Iman and Davenport’s test
This is a metric derived from Friedman’s statistic, given that this last metric produces a conservative undesirable effect. The statistic is:
and it is distributed according to a F-distribution with k − 1 and (k − 1)(N ds − 1) degrees of freedom.
3.3 Wilcoxon’s signed-rank test
This is analogous to the paired t test in non-parametrical statistical procedures; therefore, it is a pair test that aims to detect significant differences between the behaviour of two algorithms.
Let d i be the difference between the performance score of the two classifiers on ith out of N ds data-sets. The differences are ranked according to their absolute values; average ranks are assigned in case of ties. Let R + be the sum of ranks for the data-sets is which the first algorithm outperformed the second, and R − the sum of ranks for the opposite. For ranks of d i = 0 are split evenly the sums; if there is an odd number of them, one is ignored:
Let T be the smallest of the sums, T = min(R + , R −). If T is less than or equal to the value of the distribution of Wilcoxon for N ds degrees of freedom, the null hypothesis of equality of means is rejected.
Rights and permissions
About this article
Cite this article
Perez-Godoy, M.D., Rivera, A.J., Berlanga, F.J. et al. CO2RBFN: an evolutionary cooperative–competitive RBFN design algorithm for classification problems. Soft Comput 14, 953–971 (2010). https://doi.org/10.1007/s00500-009-0488-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-009-0488-z