Abstract
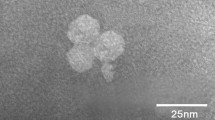
A new cyanophage, S-B05, infecting a phycoerythrin-enriched (PE-type) Synechococcus strain was isolated by the liquid infection method, and its morphology and genetic features were examined. Phylogenetic analysis and morphological observation confirmed that S-B05 belongs to the family Myoviridae of the order Caudovirales. Its genome was fully sequenced, and found to be 208,857 bp in length with a G + C content of 39.9%. It contained 280 potential open reading frames and 123 conserved domains. Ninety-eight functional genes responsible for cyanophage structuring and packaging, DNA replication and regulation, and photosynthesis were identified, as well as genes encoding 172 hypothetical proteins. The genome of S-B05 is most similar to that of Prochlorococcus phage P-TIM68. Homologues of open reading frames of S-B05 can be found in various marine environments, as revealed by comparison of the S-B05 genome sequence to sequences in marine viral metagenomic databases. The presence of auxiliary metabolic genes (AMGs) related to photosynthesis, carbon metabolism, and phosphorus assimilation, as well as the phylogenetic relationships based on AMGs and the complete genome sequence, reflect the phage-host interaction mechanism or the specific adaptation strategy of the host to environmental conditions. The genome sequence information reported here will provide an important basis for further study of the adaptive evolution and ecological role of cyanophages and their hosts in the marine environment.





Similar content being viewed by others
References
Carmichael W (1994) The toxins of cyanobacteria. Sci Am 270(1):78–86. https://www.jstor.org/stable/24942554
Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WKW, Lomas MW, Veneziano D, Vera CS, Vrugt JA, Martiny AC (2013) Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. PNAS 110(24):9824–9829. https://doi.org/10.1073/pnas.1307701110
Buitenhuis ET, Li WKW, Vaulot D, Lomas MW, Landry MR, Partensky F, Karl DM, Ulloa O, Campbell L, Jacquet S, Lantoine F, Chavez F, Macias D, Gosselin M, McManus GB (2012) Picophytoplankton biomass distribution in the global ocean. Earth Syst Sci Data 4(1):37–46. https://doi.org/10.5194/essd-4-37-2012
Xia X, Vidyarathna NK, Palenik B, Lee P, Liu H (2015) Comparison of the seasonal variations of Synechococcus Assemblage structures in estuarine waters and coastal waters of Hong Kong. Appl Environ Microbiol 81(21):7644. https://doi.org/10.1128/AEM.01895-15
Haverkamp T, Schouten D, Doeleman M et al (2009) Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the baltic sea. ISME J 3(4):397. https://doi.org/10.1038/ismej.2008.118
Suttle C, Chan A, Cottrell M (1990) Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467–469. https://doi.org/10.1038/347467a0
Suttle C (2000) Cyanophages and their role in the ecology of cyanobacteria. In: Whitton B, Potts M (eds) The ecology of cyanobacteria. Springer, Dordrecht, pp 563–589. https://doi.org/10.1007/0-306-46855-7_20
Suttle C (1994) Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol 60(9):3167–3174
Marston MF, Martiny JB (2016) Genomic diversification of marine cyanophages into stable ecotypes. Environ Microbiol 18(11):4240–4253. https://doi.org/10.1111/1462-2920.13556
Gregory A, Solonenko S, Ignacio-Espinoza J et al (2016) Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer. BMC Genom. https://doi.org/10.1186/s12864-016-3286-x
Yang Q, Gao C, Jiang Y, Wang M, Zhou X, Shao H, Gong Z, McMinn A (2019) Metagenomic characterization of the viral community of the South Scotia Ridge. Viruses 11(2):95. https://doi.org/10.3390/v11020095
Wilhelm S, Carberry M, Eldridge M et al (2006) Marine and freshwater cyanophages in a Laurentian Great Lake: evidence from infectivity assays and molecular analyses of g20 genes. Appl Environ Microbiol 72(7):4957–4963. https://doi.org/10.1128/AEM.00349-06
Wilson W, Joint I, Carr N et al (1993) Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. Strain WH7803. Appl Environ Microbiol 59(11):3736
Caroline C, Sandra K, Lauro FM (2018) Complete genome sequence of the cyanophage S-PRM1 isolated from Singapore coastal waters. Mar Genom 43:58–60. https://doi.org/10.1016/j.margen.2018.08.005
Vidaver AK, Koski RK, Etten JLV (1973) Bacteriophage φ6: a lipid-containing virus of pseudomonas phaseolicola. J Virol 11(5):799–805
Deveau H, Labrie SJ, Chopin MC et al (2006) Biodiversity and classification of lactococcal phages. Appl Environ Microbiol 72(6):4338–4346. https://doi.org/10.1128/AEM.02517-05
You S, Wang M, Jiang Y et al (2019) The genome sequence of a novel cyanophage S-B64 from the Yellow Sea, China. Curr Microbiol 76:681–686. https://doi.org/10.1007/s00284-019-01680-1
Buttimer C, Born Y, Lucid A, Loessner MJ, Fieseler L, Coffey A (2018) Erwinia amylovora phage vB_EamM_Y3 represents another lineage of hairy Myoviridae. Res Microbiol 169(9):505–514. https://doi.org/10.1016/j.resmic.2018.04.006
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964. https://doi.org/10.1093/nar/25.5.955
Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S (2017) ViPTree: the viral proteomic tree server. Bioinformatics 33:2379–2380. https://doi.org/10.1093/bioinformatics/btx157
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Lee I, Kim YO, Park SC, Chun J (2015) OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2):1100. https://doi.org/10.1099/ijsem.0.000760
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al (2007) The sorcerer II global ocean sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. https://doi.org/10.1371/journal.pbio.0050077
Hurwitz BL, Sullivan MB (2013) The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE 8:e57355. https://doi.org/10.1371/journal.pone.0057355
Xu Y, Zhang R, Wang N, Cai L, Tong Y, Sun Q, Chen F, Jiao N (2018) Novel phage–host interactions and evolution as revealed by a cyanomyovirus isolated from an estuarine environment. Environ Microbiol 20:2974–2989. https://doi.org/10.1111/1462-2920.14326
Lu L, Cai L, Jiao N et al (2017) Isolation and characterization of the first phage infecting ecologically important marine bacteria Erythrobacter. Virol J. https://doi.org/10.1186/s12985-017-0773-x
Liu Z, Wang M, Meng X et al (2017) Isolation and genome sequencing of a novel Pseudoalteromonas phage PH1. Curr Microbiol 74(2):1–7. https://doi.org/10.1007/s00284-016-1175-9
Dreher TW, Brown N, Bozarth CS, Schwartz AD, Riscoe E, Thrash C et al (2011) A freshwater cyanophage whose genome indicates close relationships to photosynthetic marine cyanomyophages. Environ Microbiol 13(7):1858–1874. https://doi.org/10.1111/j.1462-2920.2011.02502.x
Mann NH, Clokie MRJ, Millard A et al (2005) The genome of S-PM2, a "Photosynthetic" T4-Type bacteriophage that infects marine Synechococcus strains. J Bacteriol 187(9):3188–3200. https://doi.org/10.1128/JB.187.9.3188-3200.2005
Enav H, Béjà O, Mandel-Gutfreund Y (2012) Cyanophage tRNAs may have a role in cross-infectivity of oceanic Prochlorococcus and Synechococcus hosts. ISME J 6:619–628. https://doi.org/10.1038/ismej.2011.146
Sullivan MB, Huang KH, Ignacio-Espinoza JC et al (2010) Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol 12(11):3035–3056. https://doi.org/10.1111/j.1462-2920.2010.02280.x
Crummett LT, Puxty RJ, Weihe C (2016) The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 499:219–229. https://doi.org/10.1016/j.virol.2016.09.016
Mann N, Cook A, Millard A, Bailey S, Clokie M (2003) Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424:741–741. https://doi.org/10.1038/424741a
Philosof A, Battchikova N, Aro E, Beja O (2011) Marine cyanophages: tinkering with the electron transport chain. ISME J 5:1568–1570. https://doi.org/10.1038/ismej.2011.43
Thompson L, Zeng Q, Kelly L, Huang K, Singer A, Stubbe J, Chisholm S (2011) Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. PNAS 108(39):E757–E764. https://doi.org/10.1073/pnas.1102164108
Dwivedi B, Xue B, Lundin D, Edwards R, Breitbart M (2013) A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biolss. https://doi.org/10.1186/1471-2148-13-33
Hagay E, Mandel-Gutfreund Y, Béjà O (2014) Comparative metagenomics analyses reveal viral-induced shifts of host metabolism towards nucleotide biosysnthesis. Microbiome. https://doi.org/10.1186/2049-2618-2-9
Kelly L, Ding H, Huang K, Osburne M, Chisholm S (2013) Genetic diversity in cultured and wild marine cyanomyoviruses reveals phosphorus stress as a strong selective agent. ISME J 7:1827–1841. https://doi.org/10.1038/ismej.2013.58
Millard AD, Zwirglmaier K, Downey MJ et al (2009) Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ Microbiol 11(9):2370–2387. https://doi.org/10.1111/j.1462-2920.2009.01966.x
Huang S, Wang K, Jiao N (2012) Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage–host genetic exchanges. Environ Microbiol 14(2):540–558. https://doi.org/10.1111/j.1462-2920.2011.02667.x
Gao E, Ning D (2014) Advances in researches on cyanophage auxiliary metabolic genes. Microbiol China 41(8):1667–1674. https://doi.org/10.13344/j.microbiol.china.130650
Lindell D, Jaffe JD, Johnson ZI et al (2005) Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438(7064):86–89. https://doi.org/10.1038/nature04111
Lindell D, Sullivan MB, Johnson ZI et al (2004) Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci 101(30):11013–11018. https://doi.org/10.1073/pnas.0401526101
Zeidner G, Bielawski JP, Shmoish M et al (2010) Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ Microbiol 7(10):1505–1513. https://doi.org/10.1111/j.1462-2920.2005.00833.x
Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW (2006) Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. https://doi.org/10.1371/journal.pbio.0040234
Bograh A, Gingras Y, Tajmir-Riahi HA et al (1997) The effects of spermine and spermidine on the structure of photosystem II proteins in relation to inhibition of electron transport. FEBS Lett 402(1):41–44. https://doi.org/10.1016/S0014-5793(96)01453-6
Sullivan MB, Coleman ML, Weigele P et al (2005) Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3(5):e144. https://doi.org/10.1371/journal.pbio.0030144
Bryan MJ, Burroughs NJ, Spence EM et al (2008) Evidence for the intense exchange of MazG in marine cyanophages by horizontal gene transfer. PLoS ONE 3(4):e2048. https://doi.org/10.1371/journal.pone.0002048
Ignacio-Espinoza JC, Sullivan MB (2012) Phylogenomics of T4 cyanophages: lateral gene transfer in the ‘core’ and origins of host genes. Environ Microbiol 14(8):2113–2126. https://doi.org/10.1111/j.1462-2920.2012.02704.x
Li X, Sun Y, Wang X, Liu J, Wang G (2017) Research progress of new biomarker gene of phoH for bacteriophage genetic diversity. Biotechnol Bull 33(10):40–45. https://doi.org/10.13560/j.cnki.biotech.bull.1985.2017-0725
Xu J, Glibert PM, Liu H et al (2012) Nitrogen sources and rates of phytoplankton uptake in different regions of Hong Kong waters in summer. Estuaries Coasts 35(2):559–571. https://doi.org/10.1007/s12237-011-9456-9
Acknowledgements
This work was financially supported by the Natural Science Foundation of China (No. 41906126; 41976117; 41676178), and the Open Research Fund of LMB, SCSIO (LMB20091001). We sincerely thank Dr. Xia X. from SCSIO for isolating and providing the Synechococcus culture. We also thank the captain and crew of R/V Dongfanghong II, as well as the students and staff on board the cruise.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, T., Guo, C., Wang, M. et al. Isolation and complete genome sequence of a novel cyanophage, S-B05, infecting an estuarine Synechococcus strain: insights into environmental adaptation. Arch Virol 165, 1397–1407 (2020). https://doi.org/10.1007/s00705-020-04595-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04595-6