Abstract
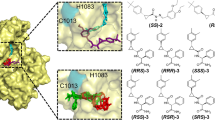
To date, no suitable vaccine or specific antiviral drug is available to treat Chikungunya viral (CHIKV) fever. Hence, it is essential to identify drug candidates that could potentially impede CHIKV infection. Here, we present the development of a homology model of nsP2 protein based on the crystal structure of the nsP2 protein of Venezuelan equine encephalitis virus (VEEV). The protein modeled was optimized using molecular dynamics simulation; the junction peptides of a nonstructural protein complex were then docked in order to investigate the possible protein–protein interactions between nsP2 and the proteins cleaved by nsP2. The modeling studies conducted shed light on the binding modes, and the critical interactions with the peptides provide insight into the chemical features needed to inhibit the CHIK virus infection. Energy-optimized pharmacophore mapping was performed using the junction peptides. Based on the results, we propose the pharmacophore features that must be present in an inhibitor of nsP2 protease. The resulting pharmacophore model contained an aromatic ring, a hydrophobic and three hydrogen-bond donor sites. Using these pharmacophore features, we screened a large public library of compounds (Asinex, Maybridge, TOSLab, Binding Database) to find a potential ligand that could inhibit the nsP2 protein. The compounds that yielded a fitness score of more than 1.0 were further subjected to Glide HTVS and Glide XP. Here, we report the best four compounds based on their docking scores; these compounds have IDs of 27943, 21362, ASN 01107557 and ASN 01541696. We propose that these compounds could bind to the active site of nsP2 protease and inhibit this enzyme. Furthermore, the backbone structural scaffolds of these four lead compounds could serve as building blocks when designing drug-like molecules for the treatment of Chikungunya viral fever.









Similar content being viewed by others
References
Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Roux KL, Prevost M-C, Fsihi H, Frenkiel M-P, Blanchet F, Afonso PV, Ceccaldi P-E, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Saib A, Rey FA, Arenzana-Seisdedos F, Despres P, Michault A, Albert ML, Schwartz O (2007) Characterization of reemerging Chikungunya virus. PLoS Pathog 3:804–817
Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney M-C, Lavenir R, Pardigon N, Reynes J-M, Pettinelli F, Biscornet L, Diancourt L, Michel S, Duquerroy S, Guigon G, Frenkiel M-P, Brhin A-C, Cubito N, Desprs P, Kunst F, Rey F, Zeller H, Brisse S (2006) Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 3:1058–1070
Ng LFP, Chow A, Sun YJ, Kwek DJC, Lim PL, Dimatatac F, Ng LC, Ooi EE, Choo KH, Her Z, Kourilsky P, Leo YS (2009) IL-β, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS ONE 4:e4261
Vanlandingham DL, Hong C, Klingler K, Tsetsarkin K, McElroy KL, Powers AM, Lehane MJ, Higgs S (2005) Differential infectivities of O’nyong-nyong and Chikungunya virus isolates in anopheles gambiae and Aedes aegypti mosquitoes. Am J Trop Med Hyg 72:616–621
Chopra A, Anuradha V, Lagoo-Joshi V, Kunjir V, Salvi S, Saluja M (2008) Chikungunya virus aches and pains: an emerging challenge. Arthritis Rheum 58:2921–2922
Pardigon N (2009) The biology of chikungunya: a brief review of what we still do not know. Pathol Biol 57:127–132
Takkinen K (1986) Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res 14:5667–5682
Khan AH, Morita K, MdC P, Hasebe F, Mathenge EGM, Igarashi A (2002) Complete nucleotide sequence of Chikungunya virus and evidence for an internal polyadenylation site. J Gen Virol 83:3075–3084
Strauss EGS, Strauss JH (1986) Structure and replication of the alphavirus genome. The Togaviridae and Flaviridae. Plenum, New York, pp 35–90
Lulla A, Lulla V, Tints K, Ahola T, Merits A (2006) Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J Virol 80:5413–5422
Perri S, Driver DA, Gardner JP, Sherrill S, Belli BA, Dubensky TW Jr, Polo JM (2000) Replicon vectors derived from Sindbis virus and Semliki Forest virus that establish persistent replication in host cells. J Virol 74:9802–9807
Zhang D, Tözsér J, Waugh DS (2009) Molecular cloning, overproduction, purification and biochemical characterization of the p39 nsp2 protease domains encoded by three alphaviruses. Protein Expr Purif 64:89–97
Vasiljeva L, Merits A, Auvinen P, Kaariainen L (2000) Identification of a novel function of the alphavirus capping apparatus. J Biol Chem 275:17281–17287
Li L, Darden T, Hiskey R, Pedersen L (1996) Homology modeling and molecular dynamics simulations of the Gla domains of human coagulation factor IX and its G[12]A mutant. J Phys Chem 100:2475–2479
Kevin JB, Edmond C, Huafeng X, Ron OD, Michael PE, Brent AG, John LK, Istvan K, Mark AM, Federico DS, John KS, Yibing S, David ES (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. ACM, Tampa
Shaw DE (2005) A fast, scalable method for the parallel evaluation of distance-limited pairwise particle interactions. J Comput Chem 26:1318–1328
Bairoch A, Boeckmann B, Ferro S, Gasteiger E (2004) Swiss-Prot: juggling between evolution and stability. Brief Bioinform 5:39–55
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Russo AT, White MA, Watowich SJ (2006) The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 14:1449–1458
Schrödinger, LLC (2009) Prime, version 2.1. Schrödinger, LLC, New York
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins Struct Funct Bioinf 17:355–362
PyMOL (2010) PyMOL molecular graphics system website. http://www.pymol.org, accessed 2010
Bowers KJ, Dror RO, Shaw DE (2006) The midpoint method for parallelization of particle simulations. J Chem Phys 124:184109–184111
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 105:6474–6487
Jørgensen AM, Topiol S (2008) Driving forces for ligand migration in the leucine transporter. Chem Biol Drug Des 72:265–272
Lauria A, Ippolito M, Almerico AM (2009) Inside the Hsp90 inhibitors binding mode through induced fit docking. J Mol Graph Model 27:712–722
Wang H, Aslanian R, Madison VS (2008) Induced-fit docking of mometasone furoate and further evidence for glucocorticoid receptor 17[alpha] pocket flexibility. J Mol Graph 27:512–521
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Salam NK, Nuti R, Sherman W (2009) Novel method for generating structure-based pharmacophores using energetic analysis. J Chem Inf Model 49:2356–2368
Nabuurs SB, Wagener M, de Vlieg J (2007) A flexible approach to induced fit docking. J Med Chem 50:6507–6518
Duffy EM, Jorgensen WL (2000) Prediction of properties from simulations: free energies of solvation in hexadecane, octanol, and water. J Am Chem 122:2878–2888
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25
Acknowledgments
The authors like to thank the Department of Bioinformatics, Alagappa University, Karaikudi, India for its support and providing the facilities for this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kh. Dhanachandra Singh, Palani Kirubakaran, and Shanthi Nagarajan contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figs. 1-4
(DOC 4739 kb)
Rights and permissions
About this article
Cite this article
Singh, K.D., Kirubakaran, P., Nagarajan, S. et al. Homology modeling, molecular dynamics, e-pharmacophore mapping and docking study of Chikungunya virus nsP2 protease. J Mol Model 18, 39–51 (2012). https://doi.org/10.1007/s00894-011-1018-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1018-3