Abstract
Single-nucleotide polymorphisms (SNPs)are molecular markers based on nucleotide variation and can be used for genotyping assays across populations and to track genomic inheritance. SNPs offer a comprehensive genotyping alternative to whole-genome sequencing for both agricultural and research purposes including molecular breeding and diagnostics, genome evolution and genetic diversity analyses, genetic mapping, and trait association studies. Here genomic SNPs were discovered between four cultivars of the important amphidiploid oilseed species Brassica napus and used to develop a B. napus Infinium™ array containing 5,306 SNPs randomly dispersed across the genome. Assay success was high, with >94 % of these producing a reproducible, polymorphic genotype in the 1,070 samples screened. Although the assay was designed to B. napus, successful SNP amplification was achieved in the B. napus progenitor species, Brassica rapa and Brassica oleracea, and to a lesser extent in the related species Brassica nigra. Phylogenetic analysis was consistent with the expected relationships between B. napus individuals. This study presents an efficient custom SNP assay development pipeline in the complex polyploid Brassica genome and demonstrates the utility of the array for high-throughput genotyping in a number of related Brassica species. It also demonstrates the utility of this assay in genotyping resistance genes on chromosome A7, which segregate amongst the 1,070 samples.



Similar content being viewed by others
References
Ahmad R, Parfitt DE, Fass J et al (2011) Whole genome sequencing of peach (Prunus persica L.) for SNP identification and selection. BMC Genomics 12:569
Akhunov E, Nicolet C, Dvorak J (2009) Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina Golden Gate assay. Theor Appl Genet 119:507–517
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anithakumari AM, Tang JF, Van Eck HJ et al (2010) A pipeline for high throughput detection and mapping of SNPs from EST databases. Mol Breed 26:65–75
Azam S, Thakur V, Ruperao P et al (2012) Coverage-based consensus calling (CBCC) of short sequence reads and comparison of CBCC results to identify SNPs in chickpea (Cicer arietinum; Fabaceae), a crop species without a reference genome. Am J Bot 99:186–192
Bachlava E, Taylor CA, Tang S et al (2012) SNP discovery and development of a high-density genotyping array for sunflower. PLoS One 7:e29814
Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS (2007) SNP discovery via 454 transcriptome sequencing. Plant J 51:910–918
Batley J, Edwards D (2007) SNP applications in plants. In: Oraguzie NC, Rikkerink EHA, Gardiner SE, De Silva HN (eds) Association mapping in plants. Springer, New York, pp 95–102
Bekele WA, Wieckhorst S, Friedt W, Snowdon RJ (2013) High-throughput genomics in sorghum: from whole-genome resequencing to a SNP screening array. Plant Biotechnol J 11(9):1112–1125
Bus A, Hecht J, Huettel B, Reinhardt R, Stich B (2012) High-throughput polymorphism detection and genotyping in Brassica napus using next-generation RAD sequencing. BMC Genomics 13:281
Cavanagh CR, Chao S, Wang S et al (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci U S A 110:8057–8062
Ccc (2010) Canola Council of Canada - Official definition of canola. http://www.canolacouncil.org/ind_definition.aspx On-line [URL] Year/Electronic Resource Number
Chagné D, Crowhurst RN, Troggio M et al (2012) Genome-wide SNP detection, validation, and development of an 8 K array for apple. PLoS One 7:e31745
Clarke WE, Parkin IA, Gajardo HA et al (2013) Genomic DNA enrichment using sequence capture microarrays: a novel approach to discover sequence nucleotide polymorphisms (SNP) in Brassica napus L. PLoS One 8
Cowling WA (2007) Genetic diversity in Australian canola and implications for crop breeding for changing future environments. Field Crop Res 104:103–111
Davey JW, Cezard T, Fuentes-Utrilla P, Eland C, Gharbi K, Blaxter ML (2013) Special features of RAD sequencing data: implications for genotyping. Mol Ecol 22:3151–3164
Delourme R, Falentin C, Fomeju BF et al (2013) High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics 14:120
Duran C, Appleby N, Vardy M, Imelfort M, Edwards D, Batley J (2009) Single nucleotide polymorphism discovery in barley using autoSNPdb. Plant Biotechnol J 7:326–333
Durstewitz G, Polley A, Plieske J et al (2010) SNP discovery by amplicon sequencing and multiplex SNP genotyping in the allopolyploid species Brassica napus. Genome 53:948–956
Eckert AJ, Van Heerwaarden J, Wegrzyn JL et al (2010) Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae). Genetics 185:969–982
Edwards D, Forster J, Chagné D, Batley J (2007) What are SNPs? In: Oraguzie N, Rikkerink E, Gardiner S, Silva H (eds) Association mapping in plants. Springer, New York, pp 41–52
Felcher KJ, Coombs JJ, Massa AN et al (2012) Integration of two diploid potato linkage maps with the potato genome sequence. PLoS One 7:e36347
Ferguson ME, Hearne SJ, Close TJ et al (2012) Identification, validation and high-throughput genotyping of transcribed gene SNPs in cassava. Theor Appl Genet 124:685–695
Friedt W, Snowdon R (2009) Oilseed rape. In: Vollmann J, Rajcan I (eds) Oil crops. Springer, New York, pp 91–126
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Report 13:207–209
Ganal MW, Durstewitz G, Polley A et al (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLOS One 6
Geraldes A, Difazio SP, Slavov GT et al (2013) A 34 K SNP genotyping array for Populus trichocarpa: design, application to the study of natural populations and transferability to other Populus species. Mol Ecol Resour 13:306–326
Hackett CA, Mclean K, Bryan GJ (2013) Linkage analysis and QTL mapping using SNP dosage data in a tetraploid potato mapping population. PLoS One 8:e63939
Hamilton JP, Hansey CN, Whitty BR et al (2011) Single nucleotide polymorphism discovery in elite North American potato germplasm. BMC Genomics 12
Haseneyer G, Schmutzer T, Seidel M et al (2011) From RNA-seq to large-scale genotyping—genomics resources for rye (Secale cereale L.). BMC Plant Biol 11:131
Hayward A, Mason A, Dalton-Morgan J, Zander M, Edwards D, Batley J (2012) SNP discovery and applications in Brassica napus. J Plant Biotechnol 39:49–61
Hu Z, Hua W, Huang S et al (2012) Discovery of pod shatter-resistant associated SNPs by deep sequencing of a representative library followed by bulk segregant analysis in rapeseed. PLoS One 7:e34253
Huang X, Wei X, Sang T et al (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Hyten DL, Song Q, Choi IY et al (2008) High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor Appl Genet 116:945–952
Hyten DL, Song QJ, Fickus EW et al (2010) High-throughput SNP discovery and assay development in common bean. BMC Genomics 11
Imelfort M, Duran C, Batley J, Edwards D (2009) Discovering genetic polymorphisms in next-generation sequencing data. Plant Biotechnol J 7:312–317
Iorizzo M, Senalik DA, Grzebelus D et al (2011) De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genomics 12
Kumar S, You FM, Cloutier S (2012) Genome wide SNP discovery in flax through next generation sequencing of reduced representation libraries. BMC Genomics 13:684
Liu S, Liu Y, Yang X et al (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930
Lorenc MT, Hayashi S, Stiller J et al (2012) Discovery of single nucleotide polymorphisms in complex genomes using SGSautoSNP. Biology 1:370–382
Myles S, Chia JM, Hurwitz B et al (2010) Rapid genomic characterization of the genus vitis. PLoS One 5(1):e8219
Novaes E, Drost DR, Farmerie WG et al (2008) High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics 9:312
Raman H, Dalton-Morgan J, Diffey S et al (2014) SNP markers-based map construction and genome-wide linkage analysis in Brassica napus. Plant Biotechnol J. doi:10.1111/pbi.12186
Rostoks N, Mudie S, Cardle L et al (2005) Genome-wide SNP discovery and linkage analysis in barley based on genes responsive to abiotic stress. Mol Gen Genomics 274:515–527
Shin JH, Blay S, Mcneney B, Graham J (2006) LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Soft 16 Code Snippet 3
Sim SC, Durstewitz G, Plieske J et al (2012a) Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS One 7(7):e40563
Sim SC, Van Deynze A, Stoffel K et al (2012b) High-density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PLoS One 7:e45520
Song Q, Hyten DL, Jia G et al (2013) Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One 8:e54985
Syvänen A-C (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet 2:930–942
Trick M, Long Y, Meng JL, Bancroft I (2009) Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J 7:334–346
Verde I, Bassil N, Scalabrin S et al (2012) Development and evaluation of a 9 K SNP array for peach by internationally coordinated SNP detection and validation in breeding germplasm. PLoS One 7
Wang XW, Wang HZ, Wang J et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–U157
Wang W, Huang S, Liu Y et al (2012) Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleracea L. var. capitata). BMC Genomics 13:523
Warnes G, Gorjanc G, Leisch F, Man M (2012) Genetics: population genetics V. R package version 1.3.7
Westermeier P, Wenzel G, Mohler V (2009) Development and evaluation of single-nucleotide polymorphism markers in allotetraploid rapeseed (Brassica napus L.). Theor Appl Genet 119:1301–1311
Wu XL, Ren CW, Joshi T, Vuong T, Xu D, Nguyen HT (2010) SNP discovery by high-throughput sequencing in soybean. BMC Genomics 11
Würschum T, Langer SM, Longin CF et al (2013) Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor Appl Genet 126:1477–1486
Yamamoto T, Nagasaki H, Yonemaru J et al (2010) Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics 11
You FM, Huo N, Deal KR et al (2011) Annotation-based genome-wide SNP discovery in the large and complex Aegilops tauschii genome using next-generation sequencing without a reference genome sequence. BMC Genomics 12:59
Acknowledgments
The authors would like to acknowledge funding support from the Australian Research Council (Projects LP0882095, LP0883462, LP0989200, LP110100200 and DP0985953). Support from the Australian Genome Research Facility (AGRF), the Queensland Cyber Infrastructure Foundation (QCIF) and the Australian Partnership for Advanced Computing (APAC) is gratefully acknowledged. Rowan Bunch is gratefully acknowledged for HiScanSQ 6 K SNP chip scanning.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Figure 1
Representative samples from the observed cluster types with Brassica napus SNPs based on the GenomeStudio software. Genotype calls are represented by different coloured regions: AA is red, BB is blue and AB is purple. (a) Locus produces well defined clusters which segregates on the A genome and fails on the C genome. (b) Locus produces well defined clusters which segregates on the C genome and fails on the A genome. (c) Locus is monomorphic within all samples run on 6 K chip. (d) Locus is monomorphic on C genome and has failed on the A genome. Individuals from diploid species are identified by the colour scheme: B. rapa: green; B. oleracea:cyan; B. nigra: pink. (GIF 334 kb)
Suppl. Figure 2
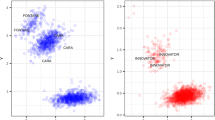
Principle component analysis on the 71 informative B. napus genotypes plus individuals from the progenitor species (46 B. rapa, four B. oleracea). (GIF 17 kb)
Suppl. Figure 3
Principle component analysis on the 71 informative B. napus genotypes. (GIF 16 kb)
Suppl. Figure 4
Heatmap displaying pairwise Linkage disequilibrium between polymorphic SNPs (PNG 1420 kb)
Table S1
(XLSX 26.6 kb)
Rights and permissions
About this article
Cite this article
Dalton-Morgan, J., Hayward, A., Alamery, S. et al. A high-throughput SNP array in the amphidiploid species Brassica napus shows diversity in resistance genes. Funct Integr Genomics 14, 643–655 (2014). https://doi.org/10.1007/s10142-014-0391-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-014-0391-2