Abstract
This study presents an integrated microfluidic system capable of rapid diagnosis of influenza infection and subtyping of influenza viruses. The viral RNA extraction module and reverse-transcription polymerase chain reaction module were integrated with an external optical detection module into the microsystem. Magnetic beads conjugated with specific nucleotide probes were first used to capture target RNA from influenza viral particles after thermolysis/hybridization processes. The hybridized target RNA was purified and collected by an external magnetic field. Finally, the extracted RNA was amplified by one-step RT-PCR products and detected by optical detection module with TaqMan fluorescence system. Moreover, the LOD was experimentally found to be 102 copies for influenza A H1/H3 and influenza B viruses. Experimental results also showed that different subtypes of influenza viruses were diagnosed by using multiplex RT-PCR process and automatically completed within 110 min in the developed microfluidic system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The most common symptoms of an influenza infection include chills, fever, sore throat, muscle pains, severe headache, coughing, weakness or fatigue and general discomfort. However, this also resulted in the deaths of between 250,000 and 500,000 people every year, and this number increase to millions in some pandemic years (Simonsen et al. 1997). On the average, 41,400 people died each year due to influenza infections in the United States between 1979 and 2001 (Dushoff et al. 2006). In April 2009, a novel influenza strain evolved from the combined genes from humans, pigs, and birds, initially dubbed as the “swine flu” and is also known as 2009 pandemic influenza A (H1N1) (infA/H1N1). It emerged in Mexico, the United States, and several other nations (Smith et al. 2009). People are generally susceptible to the 2009 new mutate of H1N1 influenza due to lack of appropriate immunity. To date, more than 10,000 cases worldwide have died of the disease. This novel influenza virus has posed a serious concern for Centers for Disease Control (CDC) around the world. Therefore, methods and tools for the rapid screening and diagnosis of seasonal and novel influenza infections are in great demand.
Influenza viruses are recognized as enveloped, segmented, negative-sense RNA viruses of the family Orthomyxoviridae and are classified into three types (A, B and C) based on the antigenic difference in their nucleoproteins (NP) and matrix proteins (M1) (Dowdle et al. 1974). Influenza A virus (infA) and B virus (infB) can cause high morbidity and mortality in humans (Matrosovich et al. 2009). InfA is one of the most virulent pathogens among the three types of influenza viruses. InfA can be further classified into a variety of subtypes according to the hemagglutinin (HA) and neuraminidase (NA) glycoproteins in the viral envelope. There are 16 subtypes of HA (H1-H16) and nine subtypes of NA (N1–N9) identified currently (Horimoto and Kawaoka 2005). Since RNA-based viruses have been characterized as having high antigenic drift and segment re-assortment, they may escape attack from the human immune system. Therefore, fast identification of influenza viruses is crucial for clinical applications.
The gold standard for diagnosis of influenza infection involves virus replication in eggs or a tissue culture followed by performing HA inhibition and NA inhibition assays (Palmer et al. 1975). However, these methods are usually time-consuming, cumbersome and technically demanding. In the past decade, some nucleic acid technology (NAT) based on a polymerase chain reaction (PCR) has been reported for the diagnosis of influenza infection with high sensitivity and high specificity (Kaul et al. 2010). There have been several laboratory assays and commercially available RT-PCR assays developed for the diagnosis of influenza viruses in the past decade (Hindiyeh et al. 2005; LeGoff et al. 2008; Smith et al. 2003). There have been several influenza assays for viral typing and subtyping developed over the last few years. Among them, the multiplex PCR has increasingly been used for the diagnosis of infectious diseases, including those caused by DNA- and RNA-based viruses (Barken et al. 2007; Elnifro et al. 2000). The major advances in multiplex RT-PCR have been made with real-time thermocyclers, or microarrays to detect infectious agents (Gall et al. 2009; Lee et al. 2008; Lin et al. 2009). However, these methods usually require bulky apparatus, experienced personnel, and lengthy process that may hinder their practical applications when pandemic infections occur.
Typically, gel electrophoresis can be used to observe the amplification of RT-PCR products. However, it usually takes about 30–60 min to separate the DNA segments according oligonucleotide length. Besides, it is not precise when quantifying the RT-PCR products. Alternatively, specific fluorophore probes allowing simultaneous amplification and visualization of the viral nucleic acids in real time have been widely used (Nazarenko 2006). For instance, the performance of the RT-PCR assays were significantly improved by using the TaqMan probe such that the sensitivity and specificity for specific gene detection was enhanced greatly (Di Trani et al. 2006). Briefly, the TaqMan probes consist of a fluorophore covalently attached to the 5′-end of the fluorescence oligonucleotide probe and a quencher at the 3′-end (Kutyavin et al. 2000). When a fluorophore and a quencher are in proximity, the quencher molecules will inhibit emission of fluorescent signals. The TaqMan probes were annealed within the amplified region. As the Taq polymerase extends the primer and synthesizes the nascent strand, the 5′–3′ exonuclease activity of the polymerase cleavages the TaqMan probe that had been annealed to the template. Degradation of the probe release the fluorophore from the 5′-end and increased the distance to the quencher, allowing the fluorescent signals to be emitted. Furthermore, it also allowed for the real-time quantification of the nucleic acids.
Recently, miniature biomedical systems integrated with functional microfluidic devices have been widely explored for molecular diagnostics. The microsystems made using micro-electro-mechanical-system (MEMS) technology have several advantages over their large-scale counterparts, including portability, lower unit cost, disposability, parallel processing, lower reagent and sample consumption, and automation (Vilkner et al. 2004). Recently, many studies have reported the detection of influenza infection by using microfluidic chips. They either used the antigen–antibody interactions to perform immunoassays or merely used purified viral RNA samples to carry out nucleic acid amplification or hybridization with specific probes on microarrays (Kao et al. 2011; Reichmuth et al. 2008). However, most devices need purified DNA or RNA prior to the diagnosis progress (Daniel et al. 2011). Furthermore, magnetic beads were extensively used in microfluidic systems for a variety of applications (Gijs 2004). For instance, specific target DNA and virus were extracted and purified efficiently with this approach (Lien et al. 2007). These magnetic beads could be further used for sample preparation and genetic amplification if integrated with on-chip PCR modules (Lien et al. 2009). Even though antibodies have been commonly used and surface-coated on magnetic beads for a variety of the applications (Lee et al. 2009; Qiu et al. 2009), there are still several critical issues when using antibodies to recognize the target proteins by magnetic beads. For example, the instability of antibodies and the cross-reactivity, which impairs diagnostic specificity, should be taken into consideration for practical applications (Magnarelli et al. 1987).
In addition, there are some commercially available microfluidic kits (for example, the FilmArray® from Idaho Technology Inc., USA) using surface-charged beads to carry out DNA extraction and purification, and then using PCR and fluorescent detection for automated rapid diagnosis of pathogens. However, the nucleic acid probe conjugated with magnetic beads has a higher specificity than surface-charged magnetic beads to capture the target samples. Alternatively, specific nucleotide probes were used to conjugate onto the surface of magnetic beads by utilizing the carboxylated linkage (Hawkins et al. 1994). The application of oligonucleotide-conjugated magnetic beads for isolation of specific nucleic acid sequences involved DNA hybridization (Jungell-Nortamo et al. 1988). Hence, sequence-specific hybridization capture on magnetic microbeads, when combined with target amplification approaches such as PCR amplification technologies, could provide a rapid and sensitive method for molecular diagnosis. It was used to capture target DNA or RNA in clinical samples (Li et al. 2008; Parham et al. 2007; Wang et al. 2007).
This study, therefore, reports an integrated microfluidic system capable of fast diagnosis and subtyping of influenza infection. It performed viral RNA extraction by using specific nucleotide probes conjugated on the surface of the magnetic beads from clinical samples by using a suction-type microfluidic control module. All the fluidic samples were mixed rapidly and manipulated precisely in an automatic manner. Moreover, multiplex RT-PCR assays for the rapid subtyping of the influenza infection, which incorporated fluorescent TaqMan probes were also implemented on the chip by utilizing the RT-PCR module with a high temperature ramping rate. Significantly, a unique, innovative feature of the suction-type, pneumatic microfluidic device for sample transportation and an efficient mixing effect was demonstrated in this study with the incorporation of a central mixing unit, a sample transportation unit and normally-closed microvalves when compared with our previous works (Lien et al. 2007, 2009). Consequently, the developed system may provide a promising platform for fast molecular diagnosis of influenza viruses.
2 Materials and methods
2.1 Working principle of the diagnostic assay
A new microsystem capable of influenza viral RNA extraction, one-step multiplex RT-PCR and optical detection of fluorescent TaqMan probes was developed in this current study. The entire process was performed automatically by carrying out a three-step operating process, namely: (1) target RNA extraction from the viral particle or clinical samples by using thermolysis and highly specific nucleotide probes-conjugated magnetic beads to capture viral RNA, (2) a one-step multiplex RT-PCR process for rapid amplification of target genes associated with subtyping of influenza viruses using highly specific subtyping primers, and (3) an end-point optical detection process by using the TaqMan probe assay. In addition, the entire diagnostic assay for diagnosis of influenza infection was performed rapidly and automatically in the suction-type microfluidic system via the on-chip mixing process and the microfluidic control module, which consist of suction-type, pneumatically driven, central mixing/transportation units. It was demonstrated that the rapid diagnosis and subtyping of influenza infection from viral or clinical sample sources with both a high sensitivity and specificity.
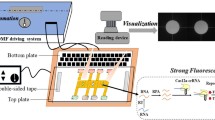
Figure 1a illustrate the working principle of the magnetic-bead-based diagnostic assay performed in the microfluidic system for the rapid identification and subtyping of influenza viruses. Figure 1b shows a photograph of the developed microfluidic chip. First, 10 μL of tested clinical samples and 5 μL of specific nucleotide probe coated magnetic beads were loaded into the clinical sample chamber. This was then followed by loading the RT-PCR reaction mixture and double-distilled water (ddH2O) (used as washing buffer in this system) into the multiplex RT-PCR chamber and the washing buffer chamber, respectively. Both the viral particles and nucleotide probe-conjugated beads were transported from clinical sample chamber to the thermal lysis/RNA hybridization chamber by using a sample transportation unit using a suction-type pump module. A temperature of 95°C was then maintained within the thermal lysis/RNA hybridization chamber to thermally lyse the influenza viruses by utilizing a temperature control module, followed by hybridizing the released viral RNA with the oligonucleotide probes-conjugated magnetic beads at 60°C. Then, a permanent magnet was attached underneath the thermal lysis/RNA hybridization chamber to collect the magnetic complexes, followed by evacuating all the other biological interference to the waste chamber with the aid of a vacuum pump. A washing process was then performed by pumping ddH2O from the washing buffer chamber to the thermal lysis/RNA hybridization chamber by using the sample transportation unit and removing the waste solution by using the permanent magnet and the vacuum pump. Thus the target viral RNA hybridized onto the magnetic beads was successfully extracted. Next, all of collected beads were re-suspended in a 25-μL solution containing an appropriate amount of ddH2O, RT-PCR reagents and TaqMan probes. All reaction mixtures were transported into the multiplex RT-PCR chamber where the one-step multiplex RT-PCR process was performed by utilizing the temperature control module. Finally, the fluorescent signals of RT-PCR amplicons were measured at end-point of the RT-PCR processes by utilizing an PMT optical detection module (Lien et al. 2007).
The working principle of the suction-type, pneumatic microfluidic control module for liquid delivery and mixing can be referenced in our previous work (Weng et al. 2011). Detail information for the chip fabrication can be found in our previous work (Weng et al. 2011). Briefly, the transport of fluidic samples was enabled when the polydimethylsiloxane (PDMS) membranes of the central mixing/transportation units were deflected upwards sequentially by negative pressure in the air chambers, generated by an external vacuum pump, such that the fluidic sample was drawn into the fluidic reservoirs underneath the central PDMS membrane. This was followed by pushing the fluidic sample from the fluidic reservoirs into the neighboring chamber when the PDMS membranes were released.
Furthermore, an efficient mixing effect was also be generated by utilizing the central mixing unit with a similar mechanism (Yang et al. 2009). For instance, two different fluidic samples were drawn simultaneously into the fluidic reservoir of the central mixing unit when the air in the air chamber of the two microvalves and the central mixing unit was evacuated with the aid of the vacuum pump. This was followed by equally distributing the sample fluids into both inlet chambers when the PDMS membrane of the central mixing unit was released. With this approach, two types of fluidic samples were rapidly mixed with a high mixing efficiency within a short period of time. All the microfluidic elements were driven by regulating electromagnetic valves (EMVs) using digital signals provided by an integrated control circuit with the incorporation of an external vacuum pump (Weng et al. 2011).
2.2 Virus preparation
The given HAU (Hemagglutination units) number from each tested influenza A (H3 and H1 subtypes) and B virus strains were titrated by a hemagglutination inhibition assay with 1% turkey erythrocytes (Donald and Isaacs 1954). The tested influenza viral isolate were confirmed, named and provided by the remaining stock samples from the National Institutes of Health (NIH) in Taiwan. In this study, the HAU of all tested virus are listed in Supplemental Table 1. All operations were carried out according to the safety instructions provided by the NIH in Taiwan.
2.3 Bioinformatic analysis of designed multiple RT-PCR primers and fluorescent probes
In this study, the nucleotide sequences of hemagglutinin were used for the diagnosis of influenza viruses by performing RT-PCR. These nucleotide sequences were selected based on sequence information obtained from the NCBI Influenza Virus Resource Database (IVRD) (http://www.ncbi.nlm.nih.gov/genomes/FLU/) (Bao et al. 2008). Ten HA types of 77 tested viral samples were used for the high consensus nucleotide regions predicted within the specific primers and probes design by using a software program, Vector NTI suite 8 (InforMax, USA). All of the tested primers, probes and TaqMan probes are presented in Table 1. The primers and TaqMan fluorescent probe specific for the hemagglutinin gene of influenza virus were synthesized from ScinoPharm® Ltd. and Protech Technology Enterprise Co., Ltd. Taiwan, respectively. The primers and probes were dissolved in sterile ddH2O and stored at −70°C prior to usage.
2.4 Viral RNA extraction protocol
The target viral RNA was captured by the specific nucleotide probe conjugated on the magnetic beads, followed by amplifying the hemagglutinin gene by one-step multiplex RT-PCR. The amplified lengths of the genes for infA/H1, infA/H3, and infB were 249, 147 and 170 base pairs (bp), respectively. Detailed information about the specific nucleotide probe conjugated onto the surface of the magnetic beads can referenced in our previous work (Wang et al. 2011). Briefly, a 10-μL tested sample of influenza virus was mixed with 5 μL of the specific nucleotide probe-conjugated magnetic beads [the diameter (Ø) of the beads 4.5 μm, concentration 4 × 107 beads/mL, MAGBEAD AGT-003-05, Applied Gene Technologies, USA] and were loaded into the thermal lysis/RNA hybridization chamber. They were then heated at 95°C for 5 min to disrupt the virus particles. Next, the reaction temperature was reduced to 60°C for 10 min so that the specific nucleotide probe could capture the target viral RNA. The entire process of viral RNA extraction was completed in approximately 15 min in an automatic manner. After the hybridization processes, 100 μL of ddH2O was loaded into the thermal lysis/RNA hybridization chamber from washing buffer chamber and pumped continuously to wash away any unbound interferents while the magnetic complexes were still restrained by a magnetic field. This washing step was repeated twice. Any cellular substances were washed away and trapped within the waste chamber by the normally-closed valve. Finally, the RT-PCR reagent with a total volume of 25 μL was loaded into the clinical sample chamber, followed by transporting them into the multiplex RT-PCR chamber.
2.5 One-step RT-PCR protocol
A one-step RT-PCR process was performed in the multiplex RT-PCR chamber. 25 μL of reaction reagents was used, which contained 10 μL of bead-captured viral RNA sample, 1 μL of deoxyribonucleotide triphosphate (dNTP, 10 mM), 2.5 μL of 10× PCR buffer (20 mM Tris–HCl, pH 8.0, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5% Tween, 0.5% Nonidet and 50% (v/v) glycerol, JMR Holdings, UK), 3 μL of multiple primers (0.5 μL of each primer for InfA/H1, InfA/H3 and InfB forward/reverse primers), 0.5 μL of Superthermo Gold Taq DNA polymerase (5 U/μL, JMR Holdings, UK) and 0.5 μL of Moloney Murine Leukemia Virus reverse transcriptase (MMLV RT, 200 U/μL, Promega, USA). Another 7.5 μL of ddH2O was finally added. The thermocycling of RT-PCR was performed with the following conditions: 42°C for 30 min (reverse transcription), 95°C for 5 min (initial denaturation), and 35 cycles of PCR at 95°C for 20 s (denaturation), 56°C for 20 s (annealing) and 72°C for 20 s (extension) for each cycle; and then at 72°C for 7 min. For the one step RT-PCR assay, the reactions were performed both in a MyCycler thermal cycler (Bio-Rad, USA) and in the integrated microfluidic system for comparison. For the negative control case, RNase-free water was used instead of the template RNA. The RT-PCR products were visualized by using gel electrophoresis on a 2% agarose with ethidium bromide staining.
2.6 Sensitivity of the one-step multiplex RT-PCR assay
The limitation of detection (LOD) for influenza virus was always determined by viral copy number in molecular diagnosis. To demonstrate the developed integrated microfluidic system has highly sensitivity in molecular diagnosis, the copies number of original tested viral samples was confirmed by real-time PCR. The tenfold series dilution of infA/H1 positive control plasmid was quantified by spectrophotometer and calculated related copy number. The real-time PCR were analyzed in triplicate with SuperScript® III Platinum® SYBR® Green One-Step qRT-PCR Kit (Invitrogen, CA, USA) by StepOnePlus real-time PCR system (Applied Biosystem, Life Technologies Co., CA, USA). Aliquots of the tested sample preparation were included in 25 μL of RT-PCR mixture containing specific primers (10 μM of each primer) and 1× one-Step RT-PCR Master Mix Reagents. Each run included a negative control (used by ddH2O) and a triplex testing. The reaction consisted of 30 min of reverse-transcription at 48°C, 10 min of activation at 95°C, and 40 cycles of amplification (95°C for 15 s and 60°C for 1 min). The standard curve was obtained by the relationship of Ct with log value about copy number of infA/H1. The RNA extracted was using PureLink™ viral RNA/DNA Mini Kit (Invitrogen, CA, USA) for three tested infA/H1 strains (128 HAU) following the previous real-time PCR condition to obtain the mean of Ct value and measured the copy number by formula. The results of the tested influenza virus isolates were analyzed for at least three tests.
2.7 Specificity of the one-step multiplex RT-PCR assay
The specificity of the one-step multiplex RT-PCR assay was determined by using three subtypes, each of them containing two or three virus isolates. The primer pairs were confirmed against the specificity panel to ensure whether the subtype-specific primer pairs were specifically recognized by the corresponding viral subtype only. The mixture contents of the PCR reagent and the protocol were the same as the ones described previously.
2.8 Optical detection of influenza viruses by the TaqMan probe
The fluorogenic TaqMan probes of infA/H1, infA/H3 and infB were labeled with 6-carboxyfluorescein (6-FAM) at the 5′ and a dark hole quencher 1 (BHQ1) dye at the 3′ end that designed in the internal region of the RT-PCR product. The TaqMan probe was designed using the Primer Express version 1.5 software (Applied Biosystems, Foster City, CA, USA). The external optical detection module was used for the rapid detection of the fluorescent signals from the amplicons of the HA genes. Light from a mercury lamp was first directed through a band-pass (BP) filter (470/20BP, Nikon Corp., Japan) and was used to excite the reporter dye (FAM) in the amplified RT-PCR products at an excitation wavelength of 488 nm. The emitted fluorescent signals from the reporter dye were then directed through a long-pass filter (505LP, Nikon Corp., Japan), followed by filtering out all other fluorescent signals excited in the PCR products utilizing another BP filter (522/16BP, Nikon Corp., Japan). Only signals with wavelengths ranging from 506 nm to 538 nm can pass through and be detected by the photomultiplier tube (PMT). With this approach, highly sensitive detection of the target genes can be achieved by utilizing the optical detection module.
As mentioned previously, the amplification of RT-PCR was monitored by using the increase in fluorescent signals that occurred when released from the TaqMan probe. The GoTaq® hot-start polymerase (Promega, USA) which exhibiting 5′ → 3′ exonuclease activity could cleave the FAM fluorphore from the TaqMan probe. The 1 μL of TaqMan probe was added in the RT-PCR reagent. The RT-PCR thermal cycling program followed the protocol described in the previous section. Statistical analyses adopting a two-tailed Student’s t test was used to determine the statistical difference. The statistical significance was confirmed when P < 0.05.
3 Results and discussion
3.1 Characterization of the microfluidic devices
The driving frequency of the PDMS membranes plays an important role in mixing for whole analytic processes. Therefore, The mixing index (σ) of the central mixing unit of the proposed microfluidic system with different driving frequencies (f d) of the PDMS membranes. The mixing indices for three different frequencies (1, 3 and 5 Hz) were investigated when the applied negative pressure was set at −80 kPa. Experimental data showed that the samples were completely mixed within 6 s when the driving frequency was 3 Hz (σ = 94.5%) (Weng et al. 2011).
Figure 2 shows the relationship between the flow-pumping rate and the applied air pressure with different diameters of the sample transportation units. Two different designs of the sample transportation unit with diameters of 5.2 and 8.0 mm were explored. A large volume of the fluidic reservoir underneath the central PDMS membrane was realized by increasing the dimensions of the PDMS membrane of the sample transportation unit, these results showed in a high flow pumping rate. A maximum flow pumping rate of approximately 444 and 650 μL/min with an air pressure of −80 kPa was achieved for units with diameters of 5.2 and 8.0 mm, respectively. Consequently, the proposed suction-type, pneumatic microfluidic device was demonstrated to be capable of handing a fluidic sample for rapid transportation and efficient mixing in an automatic manner by using the central mixing unit and the sample transportation unit when integrated with two normally-closed microvalves.
The temperature uniformity within the micro RT-PCR module was also verified at a set temperature of 95°C by an infrared (IR) imaging system (Infrared-Thermography TVS-200N, Nippon Avionics Co Ltd., Japan). The IR image showed that the microheaters with self-compensated heating grids could provide additional heating power against any thermal loss at the edges of the microheaters (Lien et al. 2009). The variation in the temperature uniformity at a set point was controlled to be within ±0.2°C. In addition, the heating and cooling rates of the module were measured to be approximately 22.3 and 13.5°C/s, respectively. Hence, these rapid heating and cooling rates assured fast nucleic acid amplification.
3.2 Specificity test
For assessment of the assay specificity, strains of infA/H1, infA/H3, infB and infA/H1N1 were tested by using the developed microfluidic system. Moreover, the target HA gene of the influenza virus particles could be rapidly lysed at 95°C and captured by the subtype-specific, probe-conjugated magnetic beads. The specificity of viral subtype for hybridization was explored and shown in Fig. 3. Using the developed microfluidic system, the infA/H1 and infA/H3 specific nucleotide probes were used for viral RNA extraction and hybridization with three infA/H1 (lanes 1–3) and two infA/H3 (lanes 4–5) strains for specificity testing, respectively. In addition, to avoid false- or cross-reactions, the infA/H1 probe-conjugated beads were incubated with the infA/H3 isolate (lane 6). Similarly, the infA/H3 probe-conjugated beads were incubated with the infA/H1 isolate (lane 7). Both of them could not generate PCR products. These results indicated that the infA/H1 or infA/H3 probe has high specificity for capturing target RNA. The two subtypes of amplified products were clearly discriminated in gel electrophoresis observation after multiplex RT-PCR (lane 8) was performed. The electropherograms showed that the two specific nucleotide probe-conjugated beads can successfully capture two subtypes of influenza viral RNA and would not interfere with each other (lane 8). This indicated that during the DNA extraction processes, various subtype-specific, probe-conjugated beads could be used together without adversely affecting the accuracy of the multiplex RT-PCR assays in this study. No cross-reactivity was observed on the specificity of the influenza A/B viruses. The experimental data, therefore, demonstrates a high specificity for the proposed one-step multiplex RT-PCR assay.
Specificity results for the nucleotide probe-conjugated magnetic beads. The infA/H1 specific probes were used for viral RNA extraction for lanes 1, 2, 3 and 6. The infA/H3 specific probes were used for viral RNA extraction for lanes 4, 5 and 7. Both infA/H1 and infA/H3 specific probes were used in lane 8. The tested infA/H1 samples were used in lanes 1, 2, 3, 7 and 8. The tested infA/H3 samples were used in lanes 4, 5, 6 and 8. L 50-bp DNA ladder markers, lane NC ddH2O was used as the template for negative control
In addition to the diagnosis of seasonal influenza, the specific primers and the nucleotide probe were designed for the rapid diagnosis of the 2009 pandemic influenza A/H1N1 virus (infA/H1N1, Supplemental Fig. 1). 64 HAU of infA/H1N1 virus were amplified by using an infA/H1N1 subtype specific probe and infA/H1N1 primer sets (lane 1). However, the results showed that lanes 2 and 3 could not be amplified by H1 and H3 specific capture probes, respectively. In contrast, the infA/H1 and infA/H3 viruses were also not identified by the infA/H1N1 specific probe (lanes 4 and 5). These results showed that the infA/H1N1 primers/probe system has a high specificity for the infA/H1N1 virus. All of these results demonstrated that the developed integrated microfluidic system could be suitable for the rapid diagnosis of various subtypes of influenza viruses. The experimental results showed that the designed probes and primers have a highly specificity for subtyping and no cross-reactions were observed with other subtypes. In addition, different subtypes of the viral isolates were added in different combinations into a multiplex RT-PCR analysis (Supplemental Fig. 2). The results showed that the system still accurately detected the desired target influenza viruses when even two (Supplemental Fig. 2, lanes 4–6) or three viruses (Supplemental Fig. 2, lane 7) subtypes coexisted in the tested sample. Therefore, several sets of primers and probes were demonstrated to apply at same time for the diagnosis and subtyping of unknown influenza viruses.
For simulating clinical examination, 106 copies of infA/H1, infA/H3 and infB viruses were added into normal nasopharyngeal swab samples to perform the microfluidic assay. RNA extraction and probe bead-based multiplex RT-PCR processes were performed according to the aforementioned method (Fig. 4). The RT-PCR products were obtained from tested viral particle added into the nasopharyngeal swab samples by using the developed integrated microfluidic system (Fig. 4a, b, c; lane 1). Furthermore, no PCR products were detected in nasopharyngeal swab samples that were not initially spiked with viruses (Fig. 4a, b, c; lane 2). This indicated that the microfluidic system has a great potential for clinical applications.
The identification of influenza viruses by using spiked clinical samples. 128 HAU of influenza A/H1 (a), A/H3 (b) and B (c) virus was added into normal nasopharyngeal swab samples to carrying out RNA extraction and probe beads-based multiplex RT-PCR processes. The specific influenza virus was added in lane 1 and nasopharyngeal swab samples were presented in lane 2. L 50-bp DNA ladder markers, lane NC ddH2O was used as the template for negative control
3.3 Sensitivity test
Following a real-time PCR assay of tested influenza virus, the detectable copy numbers of three samples (89N364H1, 89N351A and 97N510H1) with different initial concentrations of 1.2, 1.8 and 2.5 × 109 copies/μL were measured, respectively (Supplemental Fig. 3). Three infA/H1 isolates were used to explore the sensitivity and LOD of the assay. The RT-PCR results from the integrated microfluidic system were shown in Supplemental Figure 4. These tested samples were serially diluted tenfold and tested in the one-step multiplex RT-PCR assays (Supplemental Fig. 4A). The LOD of the developed system was found to be 1.2 × 102 copies in 89N364H1 (Supplemental Fig. 4B). The LOD of 89N351A and 97N510H1 was measured to be 1.8 × 101 and 2.5 × 101 copies, respectively (Supplemental Fig. 4A and 4C). Namely, the multiplex RT-PCR results showed that the microfluidic system used for the identification of influenza viruses was more sensitive by about 102–103 times than the HA inhibition assay in conventional clinical diagnostics (Gambaryan and Matrosovich 1992). Note that there are many reports or kits showing that the LOD of influenza diagnosis is about 100–101 copies by using purified viral RNA samples (Zhang and Xing 2010). However, the design of the developed microfluidic system follows the concept of lab-on-a-chip, emphasizing that sample pretreatment/purification, nucleic acid amplification and optical detection were integrated into a chip. Therefore, although the operating processes may cause the loss of small amount of RNA, the developed system still has a high LOD (102 copies/reaction).
3.4 Optical measurement of amplicons
In this study, the influenza-specific TaqMan probes were also used for optical detection of the RT-PCR amplicons. Note that the optical detection was performed for fast diagnosis after the entire process was performed on chip. Gel electrophoresis was only used for comparison. The fluorescence intensity was measured at end-point using the previously constructed optical detection apparatus in this study (Lien et al. 2009). In order to establish the optimal condition for this integrated microfluidic system, the annealing temperature, primer concentration, and RT-PCR cycling profile were considered in the previous section. The experimental fluorescent signals with a wavelength of 518 nm (with the maximum emitted fluorescent intensity) were emitted and detected by the PMT after the filter of the objective was turned on for 30 s. Note that no TaqMan probe was referenced as the negative control or a noise limit. The Rotor Gene 3000 software version 6.0.33 was used for real-time data collection and analysis.
For exploring the optimal probe concentration, three concentrations (0.1, 1, and 10 μM) of infA/H1 TaqMan probes were used for investigating the fluorescent intensity in the microfluidic system. All of the experimental results were repeated three times to calculate the average (mean) and standard deviation (SD) values. The data is shown in Table 2. The results showed that when the concentration of the TaqMan probe increased, the measured fluorescent intensity correspondingly increased. When the concentration of the TaqMan probe was 1 μM or higher, the signal-to-noise ratio (S/N) was higher than 4.9 times (341/70) which was a significant difference. The fluorescent intensity reached a maximum when the TaqMan probe concentration was 10 μM. In order to minimize the reagent consumption, 1 μM of infA/H1, infA/H3 and infB TaqMan probes was selected for the following experiments in this study.
In order to determine when the fluorescent signal from the TaqMan probe significantly increased, the variation in fluorescent intensity was measured at the end-point of different numbers of thermal cycles. These results were evaluated to identify the optimal number of thermal cycles for maximum fluorescent intensity. 1 μM of the TaqMan probes were used in 10, 20, 25 cycles of RT-PCR for fluorescent intensity testing. The final tested fluorescent intensity (∆mv) was normalized by the measured value deducted from the noise value in the end-point measurement (Table 3). The ∆mv N indicated as the final tested fluorescent intensity in the one-step multiplex RT-PCR under N cycles of amplification. The results showed that the fluorescent intensity after a 20-cycle amplification is 17.7 times (708.56/40.10) higher than the one after 10 cycles and has no obvious difference (p > 0.05) than the one after 25 cycles when using specific infA/H1 TaqMan probes. The same trend also occurred when using infA/H3 or infB TaqMan probes to detect the variation of fluorescence intensity under different PCR cycles. The fluorescent intensities of infA/H3 and infB after a 20-cycle amplification were 18.09 (609.36/17.52) and 19.03 (991.98/52.13) higher than the ones after 10 cycles and has no obvious difference with end-point detection at 25 cycles of PCR, respectively. Note that the fluorescent intensity increased with the number of thermal cycles when it was less than 20. It was speculated that the fluorescent intensity was saturated after 20 cycles by the end-point measurement (Fig. 5a). Therefore, the fluorescent intensity of ∆mv 20 was used for this analytic assay.
The fluorescent intensities for multiplex RT-PCR assays by using the specific subtype TaqMan probes to detect influenza viruses. a The effect of the number of thermal cycles on the fluorescent intensity. b The relationship between the end-point fluorescent intensity and the copy number of the target RNA for influenza A/H1, A/H3 and influenza B viruses, respectively)
According to the optimization results, 1 μM of specific TaqMan subtype probe and 20 cycles of RT-PCR were used for testing the LOD of the influenza diagnostic process. As an example, the infA/H1 samples (89N364H1) were used to perform serial tenfold dilutions in RT-PCR/TaqMan probe assays (Supplemental Table 2). Note that all three probes can be used for InfA/H1, InfA/H3 or InfB.
The correlation between different dilutions of infA/H1 and the final fluorescent intensity (∆mv 20) was further calculated and is presented by the following formula:
y = 194.52Ln(x) + 169.66 (R 2 = 0.9345)
where y represents the fluorescent intensity of ∆mv 20 and x is the diluted fold of 106 copies from tested infA/H1 strains.
Similarly, the infA/H3 and infB specific TaqMan probes were used for the LOD testing. All of the LOD for three subtypes of influenza virus were found to be 102 copies/reaction by using the developed integrated microfluidic system (Fig. 5b). The RT-PCR/TaqMan probe assay had a comparable LOD to the real-time RT-PCR/electrophoresis assay (Supplemental Fig. 3). With this approach, the fluorescent intensity of the TaqMan probe can be detected successfully and the analysis time can be shortened to 110 min.
Therefore, the TaqMan probe was used in combination with the multiplex RT-PCR for the identification of influenza viruses in the developed integrated microfluidic system. The entire process took about 110 min, which was much faster than the traditional methods that approximately needed 3–4 h. (This included commercial RNA extraction/purification, conventional RT-PCR and electrophoresis) The LOD was comparable with a real-time RT-PCR assay (Xu et al. 2008).
4 Conclusions
In this study, we demonstrated an integrated microfluidic system that combined specific probe-conjugated magnetic beads, one-step multiplex RT-PCR and a TaqMan probe assay for the diagnosis and subtyping of influenza viruses. It was capable of differentiating seasonal influenza A/H1, A/H3, pandemic influenza A/H1N1 and influenza B virus simultaneously. The integrated microfluidic system was able to perform the entire process for the detection of influenza viruses. The viral RNA extraction was automatically performed within 20 min. Several sets of highly specific probes and PCR primers were used for one-step multiplex RT-PCR. The entire process was shortening to 110 min, which was much faster than conventional methods (3–4 h). The LOD of the one-step multiplex RT-PCR microfluidic system was as low as 102 copies/reaction of influenza virus. Currently, the developed system can be used for specific type of influenza virus by performing multiplex RT-PCR with one color TaqMan probe in separate chambers. It can be easily extended for the three-color TaqMan assay if necessary. The developed microfluidic system may provide a promising platform for the fast diagnosis and subtyping of influenza infections.
References
Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D (2008) The influenza virus resource at the National Center for Biotechnology Information. J Virol 82(2):596–601
Barken KB, Haagensen JA, Tolker-Nielsen T (2007) Advances in nucleic acid-based diagnostics of bacterial infections. Clin Chim Acta 384(1–11):1–11
Daniel M, Weber P, Lutz S, Focke M, Zengerle B, von Stetten F (2011) Aliquoting on the centrigual microfluidic platform based on centrifugo-pneumatic valves. Microfluid Nanofluid 10:1279–1288
Di Trani L, Bedini B, Donatelli I, Campitelli L, Chiappini B, De Marco MA, Delogu M, Buonavolia C, Vaccari G (2006) A sensitive one-step real-time PCR for detection of avian influenza viruses using a MGB probe and an internal positive control. BMC Infect Dis 25(6):87–94
Donald HB, Isaacs A (1954) Counts of influenza virus particles. J Gen Microbiol 10(3):457–464
Dowdle WR, Galphin JC, Coleman MT, Schild GC (1974) A simple double immunodiffusion test for typing influenza viruses. Bull World Health Organ 51(3):213–215
Dushoff J, Plotkin JB, Viboud C, Earn DJ, Simonsen L (2006) Mortality due to influenza in the United States—an annualized regression approach using multiple-cause mortality data. Am J Epidemiol 163(2):181–187
Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE (2000) Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev 13(4):559–570
Gall A, Hoffman B, Harder T, Grund C, Hoper D, Beer M (2009) Design and validation of a microarray for detection, hemagglutinin subtyping, and pathotyping of avian influenza viruses. J Clin Microbiol 47(2):327–334
Gambaryan AS, Matrosovich MN (1992) A solid-phase enzyme-linked assay for influenza virus receptor-binding activity. J Virol Methods 39(1–2):111–123
Gijs MAM (2004) Magnetic bead handling on-chip: new opportunities for analytical applications. Microfluid Nanofluid 1:22–40
Hawkins TL, O’Connor-Morin T, Roy A, Santillan C (1994) DNA purification and isolation using a solid-phase. Nucleic Acids Res 22(21):4543–4544
Hindiyeh M, Levy V, Azar R, Varsano N, Regev L, Shalev Y, Grossman Z, Mendelson E (2005) Evaluation of a multiplex real-time reverse transcriptase PCR assay for detection and differentiation of influenza viruses A and B during the 2001–2002 influenza season in Israel. J Clin Microbiol 43(2):589–595
Horimoto T, Kawaoka Y (2005) Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol 3(8):591–600
Jungell-Nortamo A, Syvänen AC, Luoma P, Söderlund H (1988) Nucleic acid sandwich hybridization: enhanced reaction rate with magnetic microparticles as carriers. Mol Cell Probes 2(4):281–288
Kao LT, Shankar L, Kang TG, Zhang G, Tay GK, Rafei SR, Lee CW (2011) Multiplexed detection and differentiation of the DNA strains for influenza A (H1N1 2009) using a silicon-based microfluidic system. Biosens Bioelectron 26(5):2006–2011
Kaul KL, Mangold KA, Du H, Pesavento KM, Nawrocki J, Nowak JA (2010) Influenza A subtyping: seasonal H1N1, H3N2, and the appearance of novel H1N1. J Mol Diagn 12(5):664–669
Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J (2000) 3’-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res 28(2):655–661
Lee CS, Kang BK, Lee DH, Lyou SH, Park BK, Ann SK, Jung K, Song DS (2008) One-step multiplex RT-PCR for detection and subtyping of swine influenza H1, H3, N1, N2 viruses in clinical samples using a dual priming oligonucleotide (DPO) system. J Virol Methods 151(1):30–34
Lee YF, Lien KY, Lei HY, Lee GB (2009) An integrated microfluidic system for rapid diagnosis of dengue virus infection. Biosens Bioelectron 25(4):745–752
LeGoff J, Kara R, Moulin F, Si-Mohamed A, Krivine A, Be′lec L, Lebon P (2008) Evaluation of the one-step multiplex real-time reverse transcription-PCR ProFlu-1 assay for detection of influenza A and influenza B viruses and respiratory syncytial viruses in children. J Clin Microbiol 46(2):789–791
Li SK, Lin CH, Chen YT, Lee LH, Liu HJ (2008) Development of a reliable assay protocol for identification of diseases (RAPID)-bioactive amplification with probing for detection of avian reovirus. J Virol Methods 149(1):35–41
Lien KY, Lin JL, Liu CY, Lei HY, Lee GB (2007) Purification and enrichment of virus samples utilizing magnetic beads on a microfluidic system. Lab Chip 7(7):868–875
Lien KY, Liu CJ, Kuo PL, Lee GB (2009) Microfluidic system for detection of alpha-thalassemia-1 deletion using saliva samples. Anal Chem 81(11):4502–4509
Lin B, Malanoski AP, Wang Z, Blaney KM, Long NC, Meador CE, Metgar D, Myers CA, Yingst SL, Monteville MR, Saad MD, Schnur JM, Tibbetts C, Stenger DA (2009) Universal detection and identification of avian influenza virus by use of resequencing microarrays. J Clin Microbiol 47(4):988–993
Magnarelli LA, Anderson JF, Johnson RC (1987) Cross-reactivity in serological tests for Lyme disease and other spirochetal infections. J Infect Dis 156(1):183–188
Matrosovich M, Stech J, Klenk HD (2009) Influenza receptors, polymerase and host range. Rev Sci Tech 28(1):203–217
Nazarenko I (2006) Homogeneous detection of nucleic acids using self-quenched polymerase chain reaction primers labeled with a single fluorophore (LUX primers). Methods Mol Biol 335:95–114
Palmer DF, Dowdle WR, Coleman MT, Schild GC (1975) Advanced laboratory technicals for immunological diagnostic. Immunology Series No. 6 (US Department of Health Education and Welfare/US Public Health Service, Washington DC)
Parham NJ, Picard FJ, Peytavi R, Gagnon M, Seyrig G, Gagné PA, Boissinot M, Bergeron MG (2007) Specific magnetic bead based capture of genomic DNA from clinical samples: application to the detection of group B streptococci in vaginal/anal swabs. Clin Chem 53(9):1570–1576
Qiu J, Zhou Y, Chen H, Lin JM (2009) Immunomagnetic separation and rapid detection of bacteria using bioluminescence and microfluidics. Talanta 79(3):787–795
Reichmuth DS, Wang SK, Barrett LM, Throckmorton DJ, Einfeld W, Singh AK (2008) Rapid microchip-based electrophoretic immunoassays for the detection of swine influenza virus. Lab Chip 8(8):1319–1324
Simonsen L, Clarke MJ, Williamson GD, Stroup DF, Arden NH, Schonberger LB (1997) The impact of influenza epidemics on mortality: introducing a severity index. Am J Public Health 87(12):1944–1950
Smith AB, Mock B, Meleau R, Colarusso P, Willis DE (2003) Rapid detection of influenza A and B viruses in clinical specimens by Light Cycler real time RT-PCR. J Clin Virol 28(1):51–58
Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459(7250):1122–1125
Vilkner T, Janasek D, Manz A (2004) Micro total analysis systems. Recent developments. Anal Chem 76(12):3373–3385
Wang W, Singh S, Zeng DL, King K, Nema S (2007) Antibody structure, instability, and formulation. J Pharm Sci 96(1):1–26
Wang CH, Lien KY, Wang TY, Chen TY, Lee GB (2011) An integrated microfluidic loop-mediated-isothermal-amplification system for rapid sample pre-treatment and detection of viruses. Biosens Bioelectron 26(5):2045–2052
Weng CH, Lien KY, Yang SY, Lee GB (2011) A suction-type, pneumatic microfluidic device for liquid transport and mixing. Microfluid Nanofluid 10:301–310
Xu Y, Theobald V, Sung C, DePalma K, Atwater L, Seiger K, Perricone MA, Richards SM (2008) Validation of a HLA-A2 tetramer flow cytometric method, IFNgamma real time RT-PCR, and IFNgamma ELISPOT for detection of immunologic response to gp100 and MelanA/MART-1 in melanoma patients. J Transl Med 22:61
Yang SY, Lin JL, Lee GB (2009) A vortex-type micromixer utilizing pneumatically driven membranes. J Micromech Microeng 19. doi:10.1088/0960-1317/19/3/035020
Zhang C, Xing D (2010) Single-molecule DNA amplification and analysis using microfluidics. Chem Rev 110:4910–4947
Acknowledgments
The authors would like to thank the National Science Council in Taiwan for financial support of this project (NSC 99-2120-M-006-008). Partial financial support from “Toward A World-class University” Project is also greatly appreciated. The authors also want to acknowledge Dr. Jen-Ren Wang for kindly providing the remaining samples of the influenza A virus H1N1 from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, CH., Lien, KY., Hung, LY. et al. Integrated microfluidic system for the identification and multiple subtyping of influenza viruses by using a molecular diagnostic approach. Microfluid Nanofluid 13, 113–123 (2012). https://doi.org/10.1007/s10404-012-0947-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10404-012-0947-1