Abstract
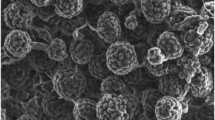
Two novel actinobacteria, designated strains NEAU-QS7T and NEAU-ML10T, were isolated from a root of Sonchus oleraceus L. and a Kronopolites svenhedind Verhoeff specimen, respectively, collected from Wuchang, Heilongjiang Province, China. A polyphasic study was carried out to establish the taxonomic positions of these strains. The two strains were observed to form abundant aerial hyphae that differentiated into spherical spore vesicles. The phylogenetic analysis based on the 16S rRNA gene sequences of strains NEAU-QS7T and NEAU-ML10T showed that the two novel isolates exhibited 99.7 % 16S rRNA gene sequence similarity with each other and that they are most closely related to Streptosporangium shengliense NEAU-GH7T (99.1, 99.0 %) and Streptosporangium longisporum DSM 43180T (99.1, 99.0 %). However, the DNA–DNA hybridization value between strains NEAU-QS7T and NEAU-ML10T was 46.5 %, and the values between the two strains and their closest phylogenetic relatives were also below 70 %. With reference to phenotypic characteristics, phylogenetic data and DNA–DNA hybridization results, the two strains can be distinguished from each other and their closest phylogenetic relatives. Thus, strains NEAU-QS7T and NEAU-ML10T represent two novel species of the genus Streptosporangium, for which the names Streptosporangium sonchi sp. nov. and Streptosporangium kronopolitis sp. nov. are proposed. The type strains are NEAU-QS7T (=CGMCC 4.7142T =DSM 46717T) and NEAU-ML10T (=CGMCC 4.7153T =DSM 46720T), respectively.


Similar content being viewed by others
References
Atlas RM (1993) Handbook of microbiological media. In: Parks LC (ed) CRC Press, Boca Raton
Collins MD (1985) Isoprenoid quinone analyses in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–284
Couch JN (1955) A new genus and family of the Actinomycetales, with a revision of the genus Actinoplanes. J Elisha Mitchell Sci Soc 71:148–155
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gao RX, Liu CX, Zhao JW, Jia FY, Yu C, Yang LY, Wang XJ, Xiang WS (2014) Micromonospora jinlongensis sp. nov., isolated from muddy soil in China and emended description of the genus micromonospora. Antonie van Leeuwenhoek 105:307–315
Goodfellow M, Stanton LJ, Simpson KE, Minnikin DE (1990) Numerical and chemical classification of Actinoplanes and some related actinomycetes. J Gen Microbiol 136:19–36
Gordon RE, Barnett DA, Handerhan JE, Pang C (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Hayakawa M, Nonomura H (1987) Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol 65:501–509
He HR, Zhang XH, Wang HY, Liu CX, Wang SR, Zhao JW, Yuan JH, Wang XJ, Xiang WS (2014) Streptosporangium nanhuense sp. nov., a novel actinomycete isolated from soil. Antonie Van Leeuwenhoek 105:1025–1031
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrometric determination of DNA hybridisation from renaturation rates. Syst Appl Microbiol 4:184–192
Intra B, Matsumoto A, Inahashi Y, Ōmura S, Panbangred W, Takahashi Y (2014) Streptosporangium jomthongense sp. nov., a novel actinomycete isolated from rhizospheric soil in Thailand and emendation of the genus Streptosporangium. Int J Syst Evol Microbiol 64:2400–2406
Jones KL (1949) Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelia is a fluctuating characteristic. J Bacteriol 57:141–145
Kelly KL (1964) Inter-society color council-national bureau of standards color-name charts illustrated with centroid colors. US Government Printing Office, Washington
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lechevalier MP, Lechevalier HA (1980) The chemotaxonomy of actinomycetes. In: Dietz A, Thayer DW (eds) Actinomycete taxonomy special publication, vol 6. Society of Industrial Microbiology, Arlington, pp 227–291
Lee YK, Kim HW, Liu CL, Lee HK (2003) A simple method for DNA extraction from marine bacteria that produce extracellular materials. J Microbiol Methods 52:245–250
Loqman S, Barka EA, Clément C, Ouhdouch Y (2009) Antagonistic actinomycetes from Moroccan soil to control the grapevine gray mold. World J Microbiol Biotechnol 25:81–91
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12B:195–206
McKerrow J, Vagg S, McKinney T, Seviour EM, Maszenan AM, Brooks P, Se-viour RJ (2000) A simple HPLC method for analysing diaminopimelic acid diastereomers in cell walls of gram-positive bacteria. Lett Appl Microbiol 30:178–182
Minnikin DE, Hutchinson IG, Caldicott AB, Goodfellow M (1980) Thin-layer chromatography of methanolysates of mycolic acid-containing bacteria. J Chromatogr 188:221–233
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int Syst Bacteriol 16:313–340
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Uchida K, Kudo T, Suzuki K, Nakase T (1999) A new rapid method of glycolate test by diethyl ether extraction, which is applicable to a small amount of bacterial cells of less than one milligram. J Gen Appl Microbiol 45:49–56
Waksman SA (1961) The actinomycetes, classification, identification and descriptions of genera and species, vol 2. Williams and Wilkins, Baltimore
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, International Committee on Systematic Bacteriology (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wu C, Lu X, Qin M, Wang Y, Ruan J (1989) Analysis of menaquinone compound in microbial cells by HPLC. Microbiology 16:176–178 [English translation of Microbiology (Beijing)]
Xiang WS, Liu CX, Wang XJ, Du J, Xi LJ, Huang Y (2011) Actinoalloteichus nanshanensis sp. nov., isolated from the rhizosphere of a fig tree (Ficus religiosa). Int J Syst Evol Microbiol 61:1165–1169
Yokota A, Tamura T, Hasegawa T, Huang LH (1993) Catenuloplanes japonicas gen. nov., sp. nov., nom. rev., a new genus of the order Actinomycetales. Int J Syst Bacteriol 43:805–812
Yu C, Liu CX, Wang XJ, Zhao JW, Yang LY, Gao RX, Zhang YJ, Xiang WS (2013) Streptomyces polyrhachii sp. nov., a novel actinomycete isolated from the edible Chinese black ant (Polyrhachis vicina Roger). Antonie Van Leeuwenhoek 104:1013–1019
Zhang XH, Liu CX, Zhang YJ, Wang HY, Wang SR, Li C, Wang XJ, Xiang WS (2014) Streptosporangium shengliensis sp. nov., a novel actinomycete isolated from a lake sediment. Antonie van Leeuwenhoek 105:237–243. Erratum to: Streptosporangium shengliensis sp. nov., a novel actinomycete isolated from a lake sediment. Antonie van Leeuwenhoek 105:265
Acknowledgments
This work was supported in part by grants from the National Outstanding Youth Foundation (No. 31225024), the National Natural Science Foundation of China (No. 31471832, 31171913 and 31372006), the National Key Technology R&D Program (No. 2012BAD19B06), the Program for New Century Excellent Talents in University (NCET-11-0953), the Outstanding Youth Foundation of Heilongjiang Province (JC201201) and Chang Jiang Scholar Candidates Program for Provincial Universities in Heilongjiang (CSCP), the Northeast Agricultural University Innovation Foundation For Postgraduate (yjscx14054), the Science and Technology Research Project of Heilongjiang Provincial Educational Commission (No. 12541001) and the Youth Science Foundation of Heilongjiang Province (No. QC2014C013). We are grateful to Prof. Aharon Oren for helpful advice on the specific epithet.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Zhaoxu Ma and Hui Liu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, Z., Liu, H., Liu, C. et al. Streptosporangium sonchi sp. nov. and Streptosporangium kronopolitis sp. nov., two novel actinobacteria isolated from a root of common sowthistle (Sonchus oleraceus L.) and a millipede (Kronopolites svenhedind Verhoeff). Antonie van Leeuwenhoek 107, 1491–1499 (2015). https://doi.org/10.1007/s10482-015-0443-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0443-1