Abstract
Two acidophilic actinobacteria, isolates NA14 and NF37T, were the subject of a polyphasic taxonomic study. Chemotaxonomic and morphological properties of the isolates were characteristic of the genus Streptacidiphilus. The isolates were shown to have identical 16S rRNA gene sequences and to be closely related to Streptacidiphilus neutrinimicus DSM 41755T (>99.9 %). However, DNA:DNA relatedness between isolate NF37T and the type strain of S. neutrinimicus was found to be low at 11.1 (±3.5) %. A broad range of phenotypic features were shown to distinguish the isolates from their close phylogenetic neighbours. These data shown that the isolates form a novel species of Streptacidiphilus for which the name Streptacidiphilus toruniensis sp. nov. is proposed. The type strain is NF37T (= DSM 102291T = NCIMB 15025T).
Similar content being viewed by others
Introduction
The genus Streptacidiphilus was proposed by Kim et al. (2003) who classified it in the family Streptomycetaceae along with the genera Kitasatospora and Streptomyces. The taxonomic status of the genera Kitasatospora and Streptacidiphilus have been questioned (Kämpfer 2012a; Labeda et al. 2012); though extensive molecular systematic data support the continued recognition of the former as a separate genus (Ishikawa et al. 2010; Girard et al. 2014), corresponding studies on the genus Streptacidiphilus are awaited.
Streptacidiphili are currently assigned to ten species with validly published names most of which have been circumscribed in extensive polyphasic studies (Golinska et al. 2013a, b). Streptacidiphilus strains grow between pH 3.5 and 6.0, optimally around pH 5.0, form extenstively branched substrate hyphae which differentiate into flexuous to straight chains of smooth-surfaced spores, contain major proportioins of LL-diaminopimelic acid in whole-organism hydrolysates, saturated, iso- and anteiso fatty acids, hexa- and octahydrogenated menaquinones with nine isoprene units as predominant isoprenologues, and complex polar lipid patterns that include diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol and phosphatidylinositol mannosides (Kämpfer 2012a, b). Streptacidiphili are common and widely distributed in acidic habitats, notably in coniferous soils (Williams et al. 1971; Golinska et al. 2013a, b), produce chitinases and diastases with low pH optima (Williams and Flowers 1978), are a source of antifungal compounds (Williams and Khan 1974) and are implicated in the turnover of organic matter at low pH values (Goodfellow and Williams 1983; Williams et al. 1984).
In a continuation of our investigations on the diversity of acidophilic sporoactinomycetes in acid forest soils, several strains were isolated that had colonial properties typical of streptacidiphili. Two of these strains, isolates NA14 and NF37T, were included in a polyphasic taxonomic study which showed that they represent a novel Streptacidophilus species for which we propose the name Streptacidiphilus toruniensis sp. nov.
Materials and methods
Sampling site
Soil samples were collected from a pine forest on the northern slope of an inland sand dune in the Torun Basin, Poland (52o55′ 37″N, 18o42′11″E) in October 2013. The trees were planted in 1875. The pine needles formed a well defined horizon which was divided into three layers; the L layer which consisted of about 3 cm of intact needles, a 3 cm deep F layer of partially decomposed but recognisable needles and a 2.5 cm deep H layer of a black-brown amorphous mass of decomposed needles, humic material and other organic matter. The underlying A1 horizon was 8 cm deep. The mean pH of the three litter layers and the mineral horizon were 4.4, 4.0, 3.7 and 3.7, respectively.
Organisms, maintenance and biomass preparation
Acidophilic filamentous sporoactinobacteria were sought from the L, F, H and A1 horizons of the pine forest soil by spreading aliquots (100 µl) of serial dilutions over the surfaces of oven dried plates of starch – casein agar (Kűster and Williams 1964) supplemented with cycloheximide and nystatin (each at 50 µg ml−1) and adjusted to pH 4.5 with 1 N HCl. The inoculated plates were incubated at 28 °C for 4 weeks, after which Streptacidiphilus-like colonies were detected from suspensions prepared from each of the environmental samples. Nine of the isolates were found to grow on starch-casein agar at pH 4.5 but not at pH 7.5.
The isolates were maintained on acidified starch-casein agar slopes at room temperature and as suspensions of mycelial fragments and spores in glycerol (20 %, v/v) at −80 °C. Strains NF37T and NA14 which were isolated from the F and A1 horizons, respectively, were chosen for detailed taxonomic analyses together with the type strains of Streptacidiphilus durhamensis, Streptacidiphilus hamsterleyensis and Streptacidiphilus neutrinimicus.
Biomass for the molecular systematic and most of the chemotaxonomic studies was prepared by cultivating the two isolates in shake flasks of acidified yeast extract-malt extract (ISP2; International Streptomyces Project medium 2, Shirling and Gottlieb 1966) broth (pH5.5), at 150 revolutions per minute at 28 °C for 2 weeks. Cells were harvested by centrifugation and washed twice in distilled water; biomass for the chemotaxonomic analyses was freeze dried and that for the molecular systematic work stored at −20 °C. Biomass (~40 mg) for the fatty acid analysis carried out on isolate NF37T was prepared by scraping growth from plates of ISP2 agar, adjusted to pH 5.5, following incubation at 28 °C for 7 days.
Phylogenetic analyses
Extraction of genomic DNA, PCR-mediated amplification of 16S rRNA genes of the two isolates and direct sequencing of the purified PCR products were carried out as described by Golinska et al. (2013c). The closest phylogenetic neighbours based on 16S rRNA gene sequence similarities were found using the EzTaxon server (http://eztaxon-e.ezbiocloud.net/, Kim et al. 2012). The resultant 16S rRNA gene sequences were aligned with those of the type strains of Streptacidiphilus species using Clustal W. Phylogenetic analyses were carried out using MEGA 6 (Tamura et al. 2013) and PHYML (Guindon and Gascuel 2003) software packages following multiple alignment using Clustal W. Evolutionary distances were calculated and clustering determined using the maximum-likelihood, maximum-parsimony and neighbour-joining methods and the resultant tree topologies evaluated by a bootstrap analysis (Felsenstein 1985) of the neighbour-joining method based on 1000 resamplings using the MEGA 6 software. The trees were rooted using the 16S rRNA gene sequence of Streptomyces albus subsp. albus DSM 40313T (GenBank accession number AJ621602).
DNA:DNA relatedness
DNA:DNA relatedness values (∆Tm) between isolate NF37T and S. neutrinimicus DSM 41755T were determined in duplicate at the DSMZ (Braunschweig, Germany). Cells were disrupted by using a Constant System TS 0.75 KW machine (IUL Instruments, Germany). DNA was isolated using a French pressure cell (Thermo Spectronic) and purified using a hydroxyapatite column, as described by Cashion et al. (1977). DNA–DNA hybridizations were carried out as described by De Ley et al. (1970), with the modifications described by Huss et al. (1983), using a model Cary 100 Bio UV/VIS spectrophotometer equipped with a Peltier-thermostatted 6 × 6 multicell changer and a temperature controller with an in situ temperature probe (Varian) at 71 °C.
Chemotaxonomy
Isolates NF37T and NA14 were examined for chemotaxonomic markers considered to be of value in the systematics of genera classified in the family Streptomycetaceae (Kämpfer 2012a). To this end, standard chromatographic methods were used to determine the isomers of diaminopimelic acid (Staneck and Roberts 1974), isoprenoid quinones (Collins 1985), polar lipids (Minnikin et al. 1984) and whole-organism sugars (Hasegawa et al. 1983), using appropriate controls. Cellular fatty acids extracted from isolate NF37T were methylated following the procedure described by Miller (1982) with minor modifications from Kuykendall et al. (1988). The fatty acid methyl esters were separated by gas chromatography (Hewlett Packard instrument 6890 N) and the resultant peaks automatically integrated, fatty acid names and percentages were determined using the standard Microbial Identification MIDI System; version 5 (Sasser 1990). The TSBA40 version of the MIDI database was used to identify the fatty acids. The G + C mol % of the DNA of the isolate was determined by HPLC following the procedure described by Tamaoka and Komagata (1984). Purified DNA prepared as described earlier was hydrolysed with P1 nuclease, the nucleotides dephosphorylated with bovine alkaline phosphatase (Mesbah et al. 1989) then analysed by HPLC (Shimadzu Corp., Japan). Lambda-DNA and three DNAs with published genome sequences representing a G + C range of 43–72 mol % were used as standards. G + C values were calculated from the ratio of deoxyguanosine and deoxythymidine, after Mesbah et al. (1989).
Cultural and morphological properties
The isolates were examined for cultural and morphological features following growth on acidified (pH 5.5) ISP media 1–7 (Shirling and Gottlieb 1966), as described previously (Golinska et al. 2013a). Hyphal and spore chain configuration were detected on acidified oatmeal agar (ISP medium 3; Shirling and Gottlieb 1966) after 14 days at 28 °C, using the coverslip technique of Kawato and Shinobu (1959). Spore arrangement and spore surface ornamentation of isolate NF37T were established by examining a gold-coated dehydrated preparation from the acidified oatmeal agar plate with a scanning electron microscope (Model 1430 VP, LEO Electron Microscopy Ltd, Cambridge, England) using the procedure described by O’Donnell et al. (1993).
Phenotypic tests
An extensive range of phenotypic tests were carried out on the isolates using media and methods described by Williams et al. (1983), albeit with acidified media. The ability of the isolates to grow at various temperatures (4, 10, 15, 20, 25, 30, 35 and 40 °C), pH values 4.0–7.5 at 0.5 pH intervals and NaCl concentrations (1, 3, 5, 7 and 10 %, w/v) were determined using acidified ISP2 agar (Shirling and Gottlieb 1966), apart from the temperature tests all of the media were incubated at 28 °C for 3 weeks. The results of these tests were compared with corresponding data for the type strains of S. durhamensis, S. hamsterleyensis and S. neutiminicus which had been acquired using the same media and methods. The enzymatic profiles of the isolates and the type strains of their nearest neighbours were acquired using API ZYM kits (Bio Mérieux) according to the manufacturer’s instructions.
Results
16S rRNA gene sequencing and DNA:DNA relatedness studies
Nearly complete 16S rRNA gene sequences of the isolates (1382–1393 nucleotides [nt]) NF37T and NA14 were determined. The strains were found to have identical sequences (Genbank accession numbers: KT933137 and KT933138, respectively) and to form a branch in the Streptacidiphilus 16S rRNA gene tree together with the type strains of Streptacidiphilus albus, Streptacidiphilus carbonis, S. durhamensis, S. hamsterleyensis and S. neutrinimicus (Fig. 1); this cluster was shown to be supported by a 97 % bootstrap value and by all of the tree-making algorithms. Isolates NA14 and NF37T were shown to be closely related to S. neutrinimicus DSM 41755T sharing a 16S rRNA gene sequence similarity with the latter of 99.9 %, a value found to correspond to 1 nt difference. The corresponding 16S rRNA gene sequence similarities between isolates NF37T and NA14 with the type strains of S. hamsterleyensis, S. durhamensis, S. albus and S. carbonis were shown to be 99.9 and 99.9 %, 98.7 and 98.7 %, 98.6 and 98.6 % and 98.3 and 98.3 %, respectively. The 16S rRNA gene sequence similarities with the type strains of the remaining Streptacidiphilus species fell within the range 95.8–96.8 %.
Neighbour-joining tree based on nearly complete 16S rRNA gene sequences (1376–1511 nucleotides) showing relationships between the isolates and between them and the type strains of Streptacidiphilus species. Asterisks indicate branches of the tree that were also found using the maximum-likelihood and maximum-parsimony tree-making algorithms. Numbers at the nodes are percentage bootstrap values based on 1000 re-sampled datasets, only values above 50 % are given. T, type strain. Bar 0.005 substitutions per nucleotide position. The root position of the tree was determined using Streptomyces albus subsp. albus DSM 40313T as the outgroup
The DNA:DNA similarity value between isolate NF37T and S. neutrinimicus DSM 41755T was shown to be 11.1 (±3.53) %, a result well below the 70 % cut-off point recommended for the assignment of closely related strains to the same genomic species (Wayne et al. 1987).
Chemotaxonomy
Isolates NA14 and NF37T were shown to have whole-organism hydrolysates rich in LL-diaminopimelic acid; galactose and rhamnose, contained major proportions of tetra-, hexa- and octahydrogenated menaquinones with nine isoprene units and in ratios of 1.0:2.5:2.4 and 1.0:6.1:11.5, respectively, and diphosphatidylglycerol, phosphatidylethanolamine (diagnostic marker), phosphatidylglycerol, phosphatidylinositol and phosphatidylinositol mannosides, as predominant polar lipids (see online Fig. S1). The cellular fatty acid profile of isolate NF37T was shown to consist of major proportions (>12 %) of anteiso-C15:0 (19.4 %), iso-C16:0 (15.4 %), C16:0 (16.4 %), anteiso-C17:0 (12.6 %), low proportions (>1.3 %) of iso-C14:0 (2.0 %), iso-C15:0 (6.9 %), iso-C16:1 H (1.4 %), iso-C17:1 ω9c (1.5 %), anteiso-C17:1 ω9c (2.1 %), iso-C17:0 (2.0 %), C17:0 cyclo (4.6 %) plus trace amounts of other components (<1 %). The genomic G + C content of isolate NF37T was 72.3 mol %.
Phenotypic properties
The isolates were found to have many phenotypic features in common, some of which distinguished them from their close phylogenetic neighbours (Table 1). The isolates, unlike the type strain of S. neutrinimicus, their nearest phylogenetic neighbour, produced a white aerial spore mass and moderate/strong yellow substrate mycelia on ISP7 medium and grew at 30 °C. In contrast, the S. neutrinimicus strain, unlike the isolates, was found to grow at pH 3.5, used L-arabitol and D-xylitol as sole carbon sources and displayed chymotrypsin activity. In addition, the isolates were found to have a greater capacity to grow on sole carbon and sole nitrogen sources than the type strains of S. durhamensis and S. hamsterleyensis. Additional phenotypic features of the isolates are cited in the species description.
Isolates NA14 and NF37T shared many properties in common but were found to produce distinctive aerial spore mass and substrate mycelial colours on some of the ISP media (Table 2), whilst only the former grew on adonitol, d-fructose and D-trehalose as sole carbon sources and on l-arginine and l-asparagine as sole nitrogen sources. In turn, isolate NF37T produced β-glucosidase and α-mannosidase (API ZYM tests).
Discussion
Filamentous sporactinomycetes which grow between pH 3.5 and 6.0 are a neglected group even though they are common in acidic habitats (Williams et al. 1971; Khan and Williams 1975; Goodfellow and Dawson 1978; Golinska et al. 2013a, b, 2015). The present study provides further evidence that the genus Streptacidiphilus is underspeciated (Lonsdale 1985; Seong et al. 1993, 1995) as isolates NA14 and NF37T were found to belong to a new Streptacidiphilus species for which the name Streptacidiphilus toruniensis sp. nov. is proposed. Comparative taxonomic surveys of streptacidiphili isolated from neglected and unexplored acidic habitats can be expected to throw additional light on the extent of the taxonomic diversity encompassed by the genus Streptacidiphilus.
Description of Streptacidiphilus toruniensis sp. nov
Streptacidiphilus toruniensis. N.L. masc. adj. toruniensis, belonging to the Polish city of Torun, the source of the isolates.
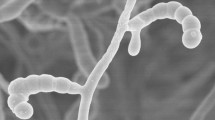
Aerobic, Gram-positive, non-acid alcohol fast, acidophilic actinobacteria which form an extensively branched substrate mycelium that carries aerial hyphae which differentiate into long straight to flexuous chains of smooth, cylindrical spores (Fig. 2) (0.6 × 0.9 µm). Grows from 10 to 30 °C, optimally ∽27 °C, from pH 4.5 to 6.5, optimally ∽pH 5.5, and in the presence of 1 %, but not at 3 % NaCl (w/v). Degrades Tweens 40 and 60, but not adenine, casein, chitin, elastin, gelatin, guanine, hypoxanthine or l-tyrosine. l-arabinose, d-cellobiose, d-galactose, d-glucosamine, d-glucose, glycogen, d-lactose, d-maltose, d-melibiose, β-methyl-D-glucoside, d-raffinose, d-rhamnose, d-salicin and d-xylose are used as sole carbon sources for energy and growth, but not d-glucuronic acid (all at 1 %, w/v). Neither acetate, adipate, benzoate, butyrate and propionate (sodium salts) nor para-hydroxybenzoic acid are used as sole carbon sources (all at 0.1 %, w/v). l-alanine, l-hydroxyproline, l-serine, and l-threonine are used as sole nitrogen sources, but not l-methionine (all at 0.1 %, w/v). Additional phenotypic properties are given in Tables 1 and 2. The major fatty acids of the type strain are anteiso-C15:0, iso-C16:0, C16:0, anteiso-C17:0 The remaining chemotaxonomic markers are typical of the genus Streptacidiphilus. The G + C content of the DNA of the type strain is 72.3 mol %.
The type strain is NF37T (= DSM 102291T = NCIMB 15025T) and strain NA14 is a second strain. These strains were isolated from the F and A1 horizons of a Pinus sylvestris L. forest in the Torun Basin, Poland.
References
Cashion P, Hodler-Franklin MA, McCully J, Franklin M (1977) A rapid method for base ratio determination of bacterial DNA. Anal Biochem 81:461–466
Collins MD (1985) Isoprenoid quinone analysis in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Girard G, Willemse J, Zhu H, Claessen D, Bukarasan K, Goodfellow M, van Wezel GP (2014) Analysis of novel kitasatosporae reveals significant evolutionary changes in conserved developmental genes between Kitasatospora and Streptomyces. Antonie Van Leeuwenhoek 106:365–380
Golinska P, Ahmed L, Wang D, Goodfellow M (2013a) Streptacidiphilus durhamensis sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek 104:199–206
Golinska P, Kim B-Y, Dahm H, Goodfellow M (2013b) Streptacidiphilus hamsterleyensis sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek 104:965–972
Golinska P, Wang D, Goodfellow M (2013c) Nocardia aciditolerans sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek 103:1079–1088
Golinska P, Zucchi TD, Silva L, Dahm H, Goodfellow M (2015) Actinospica durhamensis sp. nov., isolated from a spruce forest soil. Antonie Van Leeuwenhoek 108:435–442
Goodfellow M, Dawson D (1978) Qualitative and quantitative studies of bacteria colonizing Picea sitchensis litter. Soil Biol Bioch 10:303–307
Goodfellow M, Williams ST (1983) Ecology of actinomycetes. Ann Rev Microbiol 37:189–216
Guindon S, Gascuel O (2003) A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Ishikawa N, Oguich A, Ikeda H, Kitani S et al (2010) Genome sequence of Kitasatospora setae NBRC 14216T an evolutionary snapshot of the family Streptomycetaceae. DNA Res 17:393–406
Kämpfer P (2012a) Family I. Streptomycetaceae Waksman and Herrici 1943, 339AL emend. Rainey, Ward-Rainey and Stackebrandt 1997, 486 emend. Kim, Lonsdale, Seong and Goodfellow 2003b, 113 emend Zhi, Li and Stackebrandt 2009, 600 In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W and Whitman WB (eds) Bergey’s manual of systematic bacteriology, Second Edition, Vol 5, The Actinobacteria, Part B, Springer, New York, pp 1446–1454
Kämpfer P (2012b) Genus incertae sedis II. Streptacidiphilus Kim, Lonsdale, Seong and Goodfellow 2003a, 1219VP (Effective publication: Kim, Lonsdale, Seong and Goodfellow 2003b, 115). In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W and Whitman WB (eds) Bergey’s manual of systematic bacteriology, Second Edition, Volume 5, the Actinobacteria, Part B, Springer, New York, pp 1777–1781
Kawato M, Shinobu R (1959) On Streptomyces herbaricolor sp. nov., supplement: as simple technique for microscopical observation. Mem Osaka Univ Lib Arts Educ B Nat Sci 8:114–119
Khan MR, Williams ST (1975) Studies on the ecology of actinomycetes in soil. VIII. distribution and characteristics of acidophilic actinomycetes. Soil Biol Biochem 7:345–348
Kim SB, Lonsdale J, Seong CN, Goodfellow M (2003) Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici 1943)AL emend. Rainey et al. 1977. Antonie Van Leeuwenhoek. 83:107–116
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e, a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kűster E, Williams ST (1964) Selection of media for isolation of streptomycetes. Nature 202:928–929
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vancanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Tase A, Takahashi M, Sakane T, Suzuki K-I, Hatano K, Yamaguchi A (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104
Lonsdale JT (1985) Aspects of the biology of acidophilic actinomycetes. PhD thesis, University of Newcastle, Newcastle upon Tyne
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Miller LT (1982) A single derivatization method for bacterial fatty acid methyl esters including hydroxy acids. J Clin Microbiol 16:584–586
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
O’Donnell AG, Falconer C, Goodfellow M, Ward AC, Williams E (1993) Biosystematics and diversity amongst novel carboxydotrophic actinomycetes. Antonie Van Leeuwenhoek 64:325–340
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical note 101, MIDI Inc. Newark
Seong CN, Goodfellow M, Ward AC, Hah YC (1993) Numerical classification of acidophilic actinomycetes isolated from acid soil in Korea. Kor J Microbiol 31:355–363
Seong CN, Park SK, Goodfellow M, Kim SB, Hah YC (1995) Construction of probability identification matrix and selective medium for acidiphilic actinomycetes using numerical classification data. J Microbiol 33:95–102
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) Mega 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Wayne LG, Brenner DJ, Colwell RR et al (1987) International committee on systematic bacteriology. report on the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–465
Williams ST, Flowers TH (1978) The influence of pH on starch hydrolysis by neutrophilic and acidophilic actinomycetes. Microbios 20:99–106
Williams ST, Khan MR (1974) Antibiotics—a soil microbiologist’s viewpoint. Post Hig I Med Dosw 28:395–408
Williams ST, Davies FL, Mayfield CI, Khan MR (1971) Studies on the ecology of actinomycetes in soil. II. the pH requirements of streptomycetes from two acid soils. Soil Biol Biochem 3:187–195
Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Williams ST, Lanning S, Wellington EMH (1984) Ecology of actinomycetes. In: Goodfellow M, Mordarski M, Williams ST (eds) The biology of actinomycetes. Academic Press, London, pp 481–528
Acknowledgments
This study was supported by a Symphony 1 grant (No. 2013/08/W/NZ8/00701) from the Polish National Science Centre, and by grant from Nicolaus Copernicus University (1526B).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Golinska, P., Dahm, H. & Goodfellow, M. Streptacidiphilus toruniensis sp. nov., isolated from a pine forest soil. Antonie van Leeuwenhoek 109, 1583–1591 (2016). https://doi.org/10.1007/s10482-016-0759-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0759-5