Abstract
Objectives
To expand the repertoire of strong promoters for high level expression of proteins based on the transcriptome of Bacillus licheniformis.
Results
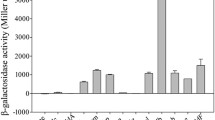
The transcriptome of B. licheniformis ATCC14580 grown to the early stationary phase was analyzed and the top 10 highly expressed genes/operons out of the 3959 genes and 1249 operons identified were chosen for study promoter activity. Using beta-galactosidase gene as a reporter, the candidate promoter pBL9 exhibited the strongest activity which was comparable to that of the widely used strong promoter p43. Furthermore, the pro-transglutaminase from Streptomyces mobaraensis (pro-MTG) was expressed under the control of promoter pBL9 and the activity of pro-MTG reached 82 U/ml after 36 h, which is 23% higher than that of promoter p43 (66.8 U/ml).
Conclusion
In our analyses of the transcriptome of B. licheniformis, we have identified a strong promoter pBL9, which could be adapted for high level expression of proteins in the host Bacillus subtilis.



Similar content being viewed by others
References
Boylan SA, Redfield AR, Price CW (1993) Transcription factor sigma B of Bacillus subtilis controls a large stationary phase regulon. J Bacteriol 175:3957–3963
Folk JE, Cole PW (1966) Identification of a functional cysteine essential for the activity of guinea pig liver transglutaminase. J Biol Chem 241:3238–3240
Georg J, Hess WR (2011) cis-antisense RNA, another level of gene regulation inbacteria. Microbiol Mol Biol Rev 75:286–300
Haldenwang WG (1995) The sigma factors of Bacillus subtilis. Microbiol Rev 59:1–30
Herman A, Halvorson H (1963) Identification of the structural gene forβ-glucosidase in Saccharomyces lactis. J Bacteriol 85:895–900
Hirata H, Negoro S, Okada H (1985) High production of thermostable β-galactosidase of Bacillus stearothermophilus in Bacillus subtilis. Appl Environ Microbiol 49:1547–1549
Jacobs M, Eliasson M, Uhlen M, Flock J (1985) Cloning, sequencing and expression of subtilisin Carlsberg from Bacillus licheniformis. Nucl Acids Res 13:8913–8926
Liao Y, Huang L, Wang B, Zhou F, Pan L (2015) The global transcriptional landscape of Bacillus amyloliquefaciens XH7 and high-throughput screening of strong promoters based on RNA-seq data. Gene 571:252–262
Martin J, Zhu W, Passalacqua KD, Bergman N, Borodovsky M (2010) Bacillus anthracis genome organization in light of whole transcriptome sequencing. BMC Bioinform 11:S10
Moreno-Campuzano S, Janga SC, Pérez-Rueda E (2006) Identification and analysis of DNA-binding transcription factors in Bacillus subtilis and other Firmicutes-agenomic approach. BMC Genom 7:147
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Nithya V, Murthy PSK, Halami PM (2013) Development and application of activefilms for food packaging using antibacterial peptide of Bacillus licheniformis Me1. J Appl Microbiol 115:475–483
Niu DD, Wang ZX (2007) Development of a pair of bifunctional expression vectors for Escherichia coli and Bacillus licheniformis. J Ind Microbiol Biotechnol 34:357–362
Pötter M, Oppermann-Sanio FB, Steinbüchel A (2001) Cultivation of bacteria producing polyamino acids with liquid manure as carbon and nitrogen source. Appl Environ Microbiol 67:617–622
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Rey MW, Ramaiya P, Nelson BA, Brody-Karpin SD, Zaretsky EJ, Tang M, de Leon AL, Xiang H, Gusti V, Clausen IG, Olsen PB, Rasmussen MD, Andersen JT, Jørgensen PL, Larsen TS, Sorokin A, Bolotin A, Lapidus A, Galleron N, Ehrlich SD, Berka RM (2004) Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol 5:R77
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17
Song Y, Nikoloff JM, Fu G, Chen J, Li Q, Xie N, Zheng P, Sun J, Zhang D (2016) Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications. PLoS ONE 11:e0158447
Sorek R, Cossart P (2010) Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11:9–16
Veith B, Herzberg C, Steckel S, Feesche J, Maurer KH, Ehrenreich P, Bäumer S, Henne A, Liesegang H, Merkl R, Ehrenreich A, Gottschalk G (2004) The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol 7:204–211
Vijayan V, Jain IH, O’Shea EK (2011) A high resolution map of a cyanobacterial transcriptome. Genome Biol 12:R47
Wang J, Ai X, Mei H, Fu Y, Chen B, Yu Z, He J (2013) High-throughput identification of promoters and screening of highly active promoter-59-UTR DNA region with different characteristics from Bacillus thuringiensis. PLoS ONE 8:e62960
Wiegand S, Dietrich S, Hertel R, Bongaerts J, Evers S, Volland S, Daniel R, Liesegang H (2013) RNA-Seq of Bacillus licheniformis: active regulatory RNA features expressed within a productive fermentation. BMC Genom 14:667
Zakataeva NP, Nikitina OV, Gronskiy SV, Romanenkov DV, Livshits VA (2010) A simple method to introduce marker-free genetic modifications into the chromosome of naturally nontransformable Bacillus amyloliquefaciens strains. Appl Microbiol Biotechnol 85:1201–1209
Acknowledgements
We wish to acknowledge the financial support by the Science and Technology Planning Project of Guangdong Province (Grant No. 2013B010404007), the State 863 Project (Grant No. 2014AA021304).
Supporting information
Supplementary Figure 1—Construction flowchart of pBE-pBL1-bgaB. All the 11 expression plasmids were constructed as follows.
Supplementary Figure 2—The homogeniety of β-Gal expression A qualitative plate assay was developed for measuring the enzyme level in isolated colonies. All materials and concentrations were the same for two plates, except for the portion of the plate that was overlaid with 3 ml of 0.067 M potassium phosphate buffer (pH 6.8) containing 3 mg of o-nitrophenyl-β-D-galactoside (ONPGal).
Supplementary Figure 3—β-Gal activities of all the promoters acting throughout the life cycle (72 h).
Supplementary Figure 4—Construction flowchart of pBE-pBL9-proMTG.
Supplementary Table 1—Bacterial strains and plasmids.
Supplementary Table 2—Primers used.
Supplementary Table 3—Genome-wide pathway analysis and GO analysis.
Supplementary Table 4—Identified operons.
Supplementary Table 5—Candidates were selected. Promoter sequence: The sequences of two operon structural genes were screened. When the sequence length was more than 300 bp, the start codon leader sequence (300 bp) was obtained. Predicted σ-factor and Transcriptional factor were obtained from: http://dbtbs.hgc.jp/.
Supplementary Table 6—Identification TF families and functional description of ten Candidates. Predicted TF families and functional description were obtained from: http://dbtbs.hgc.jp/.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
(TIFF 1495 kb)
Supplementary Figure 2
(TIFF 41,223 kb)
Supplementary Figure 3
(TIFF 1558 kb)
Supplementary Figure 4
(TIFF 1322 kb)
Supplementary Table 1
(DOCX 19 kb)
Supplementary Table 2
(DOCX 20 kb)
Supplementary Tables 3, 4, 5, 6
(XLS 1282 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Yang, H., zheng, J. et al. Identification of strong promoters based on the transcriptome of Bacillus licheniformis . Biotechnol Lett 39, 873–881 (2017). https://doi.org/10.1007/s10529-017-2304-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-017-2304-7