Abstract
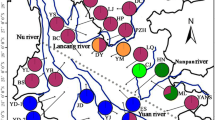
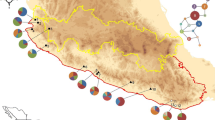
Cycas taitungensis and its sister species, C. revoluta (Cycas sect. Asiorientalis), are distributed peripatrically. Previous studies have revealed their phylogeographic pattern and, based on certain plastid DNA markers, have established that they diverged recently. However, the speciation process involving the genomic divergence has not been well clarified. In this study, based on their geographical distribution and estimates of their genetic diversity, two speciation models are proposed: (1) the founder and (2) the bottleneck speciation mode. Using approximate Bayesian computation (ABC) in combination with their estimated genetic diversity and population structure, we infer that the ancestral C. taitungensis populations diverged and southward colonized southeastern Taiwan from ancestral C. revoluta since the middle Pleistocene. These founders preserved ancestral polymorphism, resulting in small differences in genetic diversity and effective population size (Ne), despite a large difference in census population size (Nc) between two species. Based on this case of island cycad species offshore of continental Asia, this study provides new insight into how the speciation process influences the genetic diversity pattern of species with small Nc and Ne.







Similar content being viewed by others
References
Antao T, Beaumont MA (2011) Mcheza: a workbench to detect selection using dominant markers. Bioinformatics 27:1717–1718
Charlesworth B (2009) Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10:195–205
Chen M-P, Huang C-K, Lo L, Wang C-H (1992) Late Pleistocene paleoceanography of the Kuroshio Current in the area offshore Southeast Taiwan. Terr Atmos Ocean Sci 3:81–110
Chiang T-Y, Schaal BA (2006) Phylogeography of plants in Taiwan and the Ryukyu archipelago. Taxon 55:31–41
Chiang Y-C, Hung K-H, Moore S-J, Ge X-J, Huang S, Hsu T-W, Schaal BA, Chiang T (2009) Paraphyly of organelle DNAs in Cycas Sect. Asiorientales due to ancient ancestral polymorphisms. BMC Evol Bio 9:161
Chiang Y-C, Huang B-H, Liao P-C (2012) Diversification, biogeographic pattern, and demographic history of Taiwanese Scutellaria species inferred from nuclear and chloroplast DNA. PLoS ONE 7:e50844
Chiang T-Y, Chen S-F, Kato H, Hwang C-C, Moore S-J, Hsu T-W, Hung K-H (2014) Temperate origin and diversification via southward colonization in Fatsia (Araliaceae), an insular endemic genus of the West Pacific Rim. Tree Genet Genomes 10:1317–1330
Condamine FL, Nagalingum NS, Marshall CR, Morlon H (2015) Origin and diversification of living cycads: a cautionary tale on the impact of the branching process prior in Bayesian molecular dating. BMC Evol Biol 15:65
Daruka I, Ditlevsen PD (2015) A conceptual model for glacial cycles and the middle Pleistocene transition. Clim Dyn 46:29–40
Doyle J (1991) DNA protocols for plants. In: Molecular techniques in taxonomy, vol 57. Springer, Switzerland, pp 283–293
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Emerson B (2002) Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Mol Ecol 11:951–966
Frankham R (1995) Effective population size/adult population size ratios in wildlife: a review. Genet Res 66:95–107
Frankham R, Briscoe DA, Ballou JD (2010) Introduction to conservation genetics. Cambridge University Press, Cambridge
Garb JE, Gillespie RG (2009) Diversity despite dispersal: colonization history and phylogeography of Hawaiian crab spiders inferred from multilocus genetic data. Mol Ecol 18:1746–1764
Hall R (2009) Southeast Asia’s changing palaeogeography. Blumea 54:148–161
Head MJ, Gibbard PL (2015) Early-Middle Pleistocene transitions: linking terrestrial and marine realms. Quat Int 389:7–46
Hsiung HY, Huang BH, Chang JT, Huang YM, Huang CW, Liao PC (2017) Local climate heterogeneity shapes population genetic structure of two undifferentiated insular Scutellaria Species. Front Plant Sci 8:159
Hsu S-K, Sibuet J-C (1995) Is Taiwan the result of arc-continent or arc-arc collision? EPSL 136:315–324
Hu A, Meehl GA, Otto-Bliesner BL, Waelbroeck C, Han W, Loutre M-F, Lambeck K, Mitrovica JX, Rosenbloom N (2010) Influence of Bering Strait flow and North Atlantic circulation on glacial sea-level changes. Nat Geosci 3:118–121
Huang S, Chiang Y-C, Schaal BA, Chou C-H, Chiang TY (2001) Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol Ecol 10:2669–2681
Huang S, Hsieh HT, Fang K, Chiang YC (2004) Patterns of genetic variation and demography of Cycas taitungensis in Taiwan. Bot Rev 70:86–92
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Janes JK, Miller JM, Dupuis JR, Malenfant RM, Gorrell JC, Cullingham CI, Andrew RL (2017) The K = 2 conundrum. Mol Ecol 26:3594–3602
Jeffreys H (1998) The theory of probability. OUP, Oxford
Juan C, Emerson BC, Oromı́ P, Hewitt GM (2000) Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends Ecol Evol 15:104–109
Kawai K, Terada R, Kuwahara S, Takarabe M, Nishimura A, Hatta A, Hayward P, Yamamoto S, Takezaki T, Niimura H (2013) The Islands of Kagoshima: culture, society, industry and nature. Kagoshima University Research Center for the Pacific Islands, Kagoshima
Keppel G, Lee S-W, Hodgskiss P (2002) Evidence for long isolation among populations of a Pacific cycad: genetic diversity and differentiation in Cycas seemannii A. Br. (Cycadaceae). J Hered 93:133–139
Kimura M (2000) Paleogeography of the Ryukyu Islands. Tropics 10:5–24
Kimura M, Weiss GH (1964) The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics 49:561–576
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kyoda S, Setoguchi H (2010) Phylogeography of Cycas revoluta Thunb. (Cycadaceae) on the Ryukyu Islands: very low genetic diversity and geographical structure. Plant Syst Evol 288:177–189
Lariushin B (2013) Cycadaceae family, 1st edn. CreateSpace Independent Publishing Platform, Scotts Valley
Laval G, Excoffier L (2004) SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics 20:2485–2487
Luis D, Gómez P (1994) Cycads of the world. Ancient plants in today’s landscape. Econ Bot 48:34
Mayr E (1982) Speciation and macroevolution. Evolution 36:1119–1132
Nagalingum N, Marshall C, Quental T, Rai H, Little D, Mathews S (2011) Recent synchronous radiation of a living fossil. Science 334:796–799
Norstog K, Nicholls TJ (1997) The biology of the cycads. Cornell University Press, Ithaca
Osozawa S, Shinjo R, Armid A, Watanabe Y, Horiguchi T, Wakabayashi J (2012) Palaeogeographic reconstruction of the 1.55 Ma synchronous isolation of the Ryukyu Islands, Japan, and Taiwan and inflow of the Kuroshio warm current. Int Geol Rev 54:1369–1388
Ota H (1998) Geographic patterns of endemism and speciation in amphibians and reptiles of the Ryukyu Archipelago, Japan, with special reference to their paleogeographical implications. Res Popul Ecol 40:189–204
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Resour 6:288–295
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol 59:225–244
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Resour 4:137–138
Salas-Leiva DE, Meerow AW, Calonje M, Griffith MP, Francisco-Ortega J, Nakamura K, Stevenson DW, Lewis CE, Namoff S (2013) Phylogeny of the cycads based on multiple single-copy nuclear genes: congruence of concatenated parsimony, likelihood and species tree inference methods. Ann Bot 112:1263–1278
Sexton JP, Hangartner SB, Hoffmann AA (2014) Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68:1–15
Sfenthourakis S, Triantis KA (2017) The Aegean archipelago: a natural laboratory of evolution, ecology and civilisations. J Biol Res Thessalon 24:4
Sibuet J-C, Hsu S-K (1997) Geodynamics of the Taiwan arc-arc collision. Tectonophysics 274:221–251
Sibuet J-C, Hsu S-K (2004) How was Taiwan created? Tectonophysics 379:159–181
Su Y-C, Brown RM, Chang Y-H, Lin C-P, Tso IM (2016) Did a Miocene-Pliocene island isolation sequence structure diversification of funnel web spiders in the Taiwan-Ryukyu Archipelago? J Biogeogr 43:991–1003
Team RC (2015) R: a language and environment for statistical computing. https://www.R-project.org/
Templeton AR (2008) The reality and importance of founder speciation in evolution. BioEssays 30:470–479
Thieret JW (1958) Economic botany of the cycads. Econ Bot 12:3–41
Tseng H-Y, Huang W-S, Jeng M-L, Villanueva RJT, Nuñeza OM, Lin C-P (2018) Complex inter-island colonization and peripatric founder speciation promote diversification of flightless Pachyrhynchus weevils in the Taiwan-Luzon volcanic belt. J Biogeogr 45:89–100
Vos P, Hogers R, Bleeker M, Reijans M, Tvd Lee, Hornes M, Friters A, Pot J, Paleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang C, Dai J, Zhao X, Li Y, Graham SA, He D, Ran B, Meng J (2014) Outward-growth of the Tibetan Plateau during the Cenozoic: a review. Tectonophysics 621:1–43
Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L (2010) ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinform 11:116
Whitelock LM (2002) The cycads. Timber Press, Portland
Yang Y-Q, Huang B-H, Yu Z-X, Liao P-C (2015) Inferences of demographic history and fine-scale landscape genetics in Cycas panzhihuaensis and implications for its conservation. Tree Genet Genomes 11:78
Zheng Y, Liu J, Feng X, Gong X (2017) The distribution, diversity, and conservation status of Cycas in China. Ecol Evol 7:3212–3224
Acknowledgements
We would like to thank Chung-Wei You for his helpful sampling of C. revoluta among the Ryukyu Islands. We thank the National Center for Genome Medicine of the National Core Facility Program for Biotechnology, Ministry of Science and Technology, Taiwan, for technical and bioinformatics support. We acknowledge Chih-Chieh Yu and Hsiao-Lei Liu for the suggestion of contents. We also thank Dr. Hubert Turner for English editing of the manuscript. This research was financially supported by the Ministry of Science and Technology of Taiwan (Grant Number: MOST 105–2628-B-003–001-MY3 and MOST 105–2628-B-003–002-MY3) and was also subsidized by National Taiwan Normal University (NTNU), Taiwan.
Author information
Authors and Affiliations
Contributions
JTC and PCL conceived the project and collected field samples. JTC conducted genetic experiments, performed statistical analyses, interpreted the data, and wrote the manuscript. BHH assisted in the genetic diversity and evolutionary analyses. PCL critically reviewed the draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10592_2019_1193_MOESM1_ESM.tif
Supplementary material 1 (TIFF 93 kb) Supplementary Fig. 1 Isolation by distance (IBD) result by Mantel test. The pairwise correlation between genetic and geographical distance shows the non-significant result.
10592_2019_1193_MOESM3_ESM.tif
Supplementary material 3 (TIFF 689 kb) Supplementary Fig. 3 The population structure analysis from K = 5 to K = 10 with 1 million MCMC times and 10% burn-in.
10592_2019_1193_MOESM4_ESM.docx
Supplementary material 4 (DOCX 21 kb) Supplementary Table 1. The prior setting of two evolutionary scenarios. A) Bottleneck hypothesis B) Founder hypothesis. Ne, rel, m, GAMMA and rec refer to the effective population size, relative rate, migration rate, gamma distribution (for estimating recombination and mutation rate) and recombination rate.
Rights and permissions
About this article
Cite this article
Chang, JT., Huang, BH. & Liao, PC. Genetic evidence of the southward founder speciation of Cycas taitungensis from ancestral C. revoluta along the Ryukyu Archipelagos. Conserv Genet 20, 1045–1056 (2019). https://doi.org/10.1007/s10592-019-01193-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01193-1