Summary
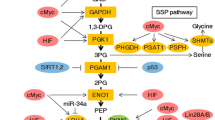
Serine and glycine are the primary sources of one-carbon units that are vital for cell proliferation. Their abnormal metabolism is known to be associated with cancer progression. As the key enzyme of serine metabolism, Serine Hydroxymethyltransferase 2 (SHMT2) has been a research hotspot in recent years. SHMT2 is a PLP-dependent tetrameric enzyme that catalyzes the reversible transition from serine to glycine, thus promoting the production of one-carbon units that are indispensable for cell growth and regulation of the redox and epigenetic states of cells. Under a hypoxic environment, SHMT2 can be upregulated and could promote the generation of nicotinamide adenine dinucleotide phosphate (NADPH) and glutathione for maintaining the redox balance. Accumulating evidence confirmed that SHMT2 facilitates cell proliferation and tumor growth and is tightly associated with poor prognosis. In this review, we present insights into the function and research development of SHMT2 and summarize the possible molecular mechanisms of SHMT2 in promoting tumor growth, in the hope that it could provide clues to more effective clinical treatment of cancer.


Similar content being viewed by others
References
Warburg O (1956) On the origin of cancer cells. Science 123(3191):309–314. https://doi.org/10.1126/science.123.3191.309
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324(5930):1029–1033. https://doi.org/10.1126/science.1160809
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer Metabolism. Cell Metab 23(1):27–47. https://doi.org/10.1016/j.cmet.2015.12.006
Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G (2014) Serine and glycine metabolism in cancer. Trends Biochem Sci 39(4):191–198. https://doi.org/10.1016/j.tibs.2014.02.004
Locasale JW (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13(8):572–583. https://doi.org/10.1038/nrc3557
Kalhan SC, Hanson RW (2012) Resurgence of serine: an often neglected but indispensable amino Acid. J Biol Chem 287(24):19786–19791. https://doi.org/10.1074/jbc.r112.357194
Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD (2014) Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep 7(4):1248–1258. https://doi.org/10.1016/j.celrep.2014.04.045
Wu XY, Lu L (2012) Vitamin B6 deficiency, genome instability and cancer. Asian Pac J Cancer Prev 13(11):5333–5338. https://doi.org/10.7314/apjcp.2012.13.11.5333
Anderson DD, Stover PJ (2009) SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS One 4(6):e5839. https://doi.org/10.1371/journal.pone.0005839
Giardina G, Brunotti P, Fiascarelli A, Cicalini A, Costa MG et al (2015) How pyridoxal 5’-phosphate differentially regulates human cytosolic and mitochondrial serine hydroxymethyltransferase oligomeric state. FEBS J 282(7):1225–1241. https://doi.org/10.1111/febs.13211
Hebbring SJ, Chai Y, Ji Y, Abo RP, Jenkins GD et al (2012) Serine hydroxymethyltransferase 1 and 2: gene sequence variation and functional genomic characterization. J Neurochem 120(6):881–890. https://doi.org/10.1111/j.1471-4159.2012.07646.x
MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, Stover PJ (2008) Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem 283(38):25846–25853. https://doi.org/10.1074/jbc.m802671200
Garrow TA, Brenner AA, Whitehead VM, Chen XN, Duncan RG, Korenberg JR, Shane B (1993) Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J Biol Chem 268(16):11910–11916
Dou C, Xu Q, Liu J, Wang Y, Zhou Z, Yao W, Jiang K, Cheng J, Zhang C (2019) Tu K (2019) SHMT1 inhibits the metastasis of HCC by repressing NOX1-mediated ROS production. J Exp Clin Cancer Res 38(1):70. https://doi.org/10.1186/s13046-019-1067-5
Woo CC, Chen WC, Teo XQ, Radda GK, Lee PT (2016) Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget 7(33):53005–53017. https://doi.org/10.18632/oncotarget.10415
Anderson DD, Quintero CM, Stover PJ (2011) Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci U S A 108(37):15163–15168. https://doi.org/10.1073/pnas.1103623108
Giardina G, Paone A, Tramonti A, Lucchi R, Marani M et al (2018) The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells. FEBS J 285(17):3238–3253. https://doi.org/10.1111/febs.14610
Cuyàs E, Fernández-Arroyo S, Verdura S, García RÁ, Stursa J et al (2018) Metformin regulates global DNA methylation via mitochondrial one-carbon metabolism. Oncogene 37(7):963–970. https://doi.org/10.1038/onc.2017.367
Parsa S, Ortega-Molina A, Ying HY, Jiang M, Teater M et al (2020) The serine hydroxymethyltransferase-2 (SHMT2) initiates lymphoma development through epigenetic tumor suppressor silencing. Nat Cancer 1:653–664. https://doi.org/10.1038/s43018-020-0080-0
Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD (2016) Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab 23(6):1140–1153. https://doi.org/10.1016/j.cmet.2016.04.016
Minton DR, Nam M, McLaughlin DJ, Shin J, Bayraktar EC et al (2018) Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol Cell 69(4):610–621.e5. https://doi.org/10.1016/j.molcel.2018.01.024
Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, Rabinowitz JD (2018) Mitochondrial translation requires folate-dependent tRNA methylation. Nature 554(7690):128–132. https://doi.org/10.1038/nature25460
Ducker GS, Rabinowitz JD (2017) One-Carbon Metabolism in Health and Disease. Cell Metab 25(1):27–42. https://doi.org/10.1016/j.cmet.2016.08.009
Hashizume O, Ohnishi S, Mito T, Shimizu A, Ishikawa K et al (2015) Epigenetic regulation of the nuclear-coded GCAT and SHMT2 genes confers human age-associated mitochondrial respiration defects. Sci Rep 5:10434. https://doi.org/10.1038/srep10434
Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW (1999) Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res 59(22):5830–5835
Kabakov AE, Yakimova AO (2021) Hypoxia-induced cancer cell responses driving radioresistance of hypoxic tumors: approaches to targeting and radiosensitizing. Cancers (Basel) 13(5):1102. https://doi.org/10.3390/cancers13051102
Graham K, Unger E (2018) Overcoming tumor hypoxia as a barrier to radiotherapy, chemotherapy and immunotherapy in cancer treatment. Int J Nanomedicine 13:6049–6058. https://doi.org/10.2147/ijn.s140462
Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H (2019) HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci 20(2):238. https://doi.org/10.3390/ijms20020238
Ye J, Fan J, Venneti S, Wan YW, Pawel BR et al (2014) Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov 4(12):1406–1417. https://doi.org/10.1158/2159-8290.CD-14-0250
Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD (2014) Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510(7504):298–302. https://doi.org/10.1038/nature13236
Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD (2002) A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol 22(16):5793–5800. https://doi.org/10.1128/mcb.22.16.5793-5800.2002
Haggerty TJ, Zeller KI, Osthus RC, Wonsey DR, Dang CV (2003) A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc Natl Acad Sci U S A 100(9):5313–5318. https://doi.org/10.1073/pnas.0931346100
Yang F, Zhang Y, Ren H, Wang J, Shang L et al (2019) Ischemia reperfusion injury promotes recurrence of hepatocellular carcinoma in fatty liver via ALOX12-12HETE-GPR31 signaling axis. J Exp Clin Cancer Res 38(1):489. https://doi.org/10.1186/s13046-019-1480-9
Wang M, Yuan F, Bai H, Zhang J, Wu H, Zheng K, Zhang W, Miao M, Gong J (2019) SHMT2 promotes liver regeneration through glycine-activated Akt/mTOR pathway. Transplantation 103(7):e188–e197. https://doi.org/10.1097/tp.0000000000002747
Maeno H, Ono T, Dhar DK, Sato T, Yamanoi A, Nagasue N (2005) Expression of hypoxia inducible factor-1alpha during liver regeneration induced by partial hepatectomy in rats. Liver Int 25(5):1002–1009. https://doi.org/10.1111/j.1478-3231.2005.01144.x
Wu H, Bai H, Duan S, Yuan F (2019) Downregulating serine hydroxymethyltransferase 2 deteriorates hepatic ischemia-reperfusion injury through ROS/JNK/ P53 signaling in mice. Biomed Res Int 2019:2712185. https://doi.org/10.1155/2019/2712185
Leivonen SK, Rokka A, Ostling P, Kohonen P, Corthals GL, Kallioniemi O, Perälä M (2011) Identification of miR-193b targets in breast cancer cells and systems biological analysis of their functional impact. Mol Cell Proteomics 10(7):M110.005322. https://doi.org/10.1074/mcp.m110.005322
Lee GY, Haverty PM, Li L, Kljavin NM, Bourgon R et al (2014) Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res 74(11):3114–3126
Zhao LN, Björklund M, Caldez MJ, Zheng J, Kaldis P (2021) Therapeutic targeting of the mitochondrial one-carbon pathway: perspectives, pitfalls, and potential. Oncogene 40(13):2339–2354. https://doi.org/10.1038/s41388-021-01695-8
Koseki J, Konno M, Asai A, Colvin H, Kawamoto K et al (2018) Enzymes of the one-carbon folate metabolism as anticancer targets predicted by survival rate analysis. Sci Rep 8(1):303. https://doi.org/10.1038/s41598-017-18456-x
Shi H, Fang X, Li Y, Zhang Y (2019) High Expression of Serine Hydroxymethyltransferase 2 Indicates Poor Prognosis of Gastric Cancer Patients. Med Sci Monit 25:7430–7438. https://doi.org/10.12659/msm.917435
Liu Y, Yin C, Deng MM, Wang Q, He XQ, Li MT, Li CP, Wu H (2019) High expression of SHMT2 is correlated with tumor progression and predicts poor prognosis in gastrointestinal tumors. Eur Rev Med Pharmacol Sci 23(21):9379–9392. https://doi.org/10.26355/eurrev
Miyo M, Konno M, Colvin H, Nishida N, Koseki J et al (2017) The importance of mitochondrial folate enzymes in human colorectal cancer. Oncol Rep 37(1):417–425. https://doi.org/10.3892/or.2016.5264
Lin C, Zhang Y, Chen Y, Bai Y, Zhang Y (2019) Long noncoding RNA LINC01234 promotes serine hydroxymethyltransferase 2 expression and proliferation by competitively binding miR-642a-5p in colon cancer. Cell Death Dis 10(2):137. https://doi.org/10.1038/s41419-019-1352-4
Ji L, Tang Y, Pang X, Zhang Y (2019) Increased expression of serine Hydroxymethyltransferase 2 (SHMT2) is a negative prognostic marker in patients with hepatocellular carcinoma and is Associated with Proliferation of HepG2 Cells. Med Sci Monit 25:5823–5832. https://doi.org/10.12659/msm.915754
Wu X, Deng L, Tang D, Ying G, Yao X, Liu F, Liang G (2016) miR-615-5p prevents proliferation and migration through negatively regulating serine hydromethyltransferase 2 (SHMT2) in hepatocellular carcinoma. Tumour Biol 37(5):6813–6821. https://doi.org/10.1007/s13277-015-4506-8
Zhang L, Chen Z, Xue D, Zhang Q, Liu X et al (2016) Prognostic and therapeutic value of mitochondrial serine hydroxylmethyltransferase 2 as a breast cancer biomarker. Oncol Rep 36(5):2489–2500. https://doi.org/10.3892/or.2016.5112
Yin K (2015) Positive correlation between expression level of mitochondrial serine hydroxylmethyltransferase and breast cancer grade. Onco Targets Ther 8:1069–1074. https://doi.org/10.2147/ott.s82433
Wang CY, Chiao CC, Phan NN, Li CY, Sun ZD et al (2020) Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am J Cancer Res 10(1):95–113
Bernhardt S, BayerlováM VM, Wachter A, Mitra D et al (2017) Proteomic profiling of breast cancer metabolism identifies SHMT2 and ASCT2 as prognostic factors. Breast Cancer Res 19(1):112. https://doi.org/10.1186/s13058-017-0905-7
Li AM, Ducker GS, Li Y, Seoane JA, Xiao Y et al (2020) Metabolic profiling reveals a dependency of human metastatic breast cancer on mitochondrial serine and one-carbon unit metabolism. Mol Cancer Res 18(4):599–611. https://doi.org/10.1158/1541-7786.mcr-19-0606
Dong Y, Huo X, Sun R, Liu Z, Huang M, Yang S (2018) lncRNA Gm15290 promotes cell proliferation and invasion in lung cancer through directly interacting with and suppressing the tumor suppressor miR-615-5p. Biosci Rep 38(5):BSR20181150. https://doi.org/10.1042/BSR20181150
DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z et al (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47(12):1475–1481. https://doi.org/10.1038/ng.3421
Engel AL, Lorenz NI, Klann K, Münch C, Depner C, Steinbach JP, Ronellenfitsch MW, Luger AL (2020) Serine-dependent redox homeostasis regulates glioblastoma cell survival. Br J Cancer 122(9):1391–1398. https://doi.org/10.1038/s41416-020-0794-x
Wang B, Wang W, Zhu Z, Zhang X, Tang F, Wang D, Liu X, Yan X, Zhuang H (2017) Mitochondrial serine hydroxymethyltransferase 2 is a potential diagnostic and prognostic biomarker for human glioma. Clin Neurol Neurosurg 154:28–33. https://doi.org/10.1016/j.clineuro.2017.01.005
Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K et al (2015) SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature 520(7547):363–367. https://doi.org/10.1038/nature14363
Liao Y, Wang F, Zhang Y, Cai H, Song F, Hou J (2021) Silencing SHMT2 inhibits the progression of tongue squamous cell carcinoma through cell cycle regulation. Cancer Cell Int 21(1):220. https://doi.org/10.1186/s12935-021-01880-5
Rabl J, Bunker RD, Schenk AD, Cavadini S, Gill ME et al (2019) Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation. Mol Cell 75(3):483-497.e9. https://doi.org/10.1016/j.molcel.2019.06.002
Rabl J (2020) BRCA1-A and BRISC: Multifunctional Molecular Machines for Ubiquitin Signaling. Biomolecules 10(11):1503. https://doi.org/10.3390/biom10111503
Walden M, Tian L, Ross RL, Sykora UM, Byrne DP, Hesketh EL, Masandi SK, Cassel J, George R et al (2019) Metabolic control of BRISC-SHMT2 assembly regulates immune signalling. Nature 570(7760):194–199. https://doi.org/10.1038/s41586-019-1232-1
Zheng H, Gupta V, Patterson-Fortin J, Bhattacharya S, Katlinski K, Wu J, Varghese B, Carbone CJ, Aressy B, Fuchs SY, Greenberg RA (2013) A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep 5(1):180–193. https://doi.org/10.1016/j.celrep.2013.08.025
Xu M, Moresco JJ, Chang M, Mukim A, Smith D, Diedrich JK, Yates JR, Jones KA (2018) SHMT2 and the BRCC36/BRISC deubiquitinase regulate HIV-1 Tat K63-ubiquitylation and destruction by autophagy. PLoS Pathog 14(5):e1007071. https://doi.org/10.1371/journal.ppat.1007071
Liu C, Wang L, Liu X, Tan Y, Tao L et al (2021) Cytoplasmic SHMT2 drives the progression and metastasis of colorectal cancer by inhibiting β-catenin degradation. Theranostics 11(6):2966–2986. https://doi.org/10.7150/thno.48699
Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL et al (2013) SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23(4):450–463. https://doi.org/10.1016/j.ccr.2013.02.024
Wang YS, Du L, Liang X, Meng P, Bi L, Wang YL, Wang C, Tang B (2019) Sirtuin 4 depletion promotes hepatocellular carcinoma tumorigenesis through regulating adenosine-monophosphate-activated protein kinase alpha/mammalian target of rapamycin axis in mice. Hepatology 69(4):1614–1631. https://doi.org/10.1002/hep.30421
Chang L, Xi L, Liu Y, Liu R, Wu Z, Jian Z (2018) SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol Med Rep 17(1):342–349. https://doi.org/10.3892/mmr.2017.7875
Ma Y, Qi Y, Wang L, Zheng Z, Zhang Y, Zheng J (2019) SIRT5-mediated SDHA desuccinylation promotes clear cell renal cell carcinoma tumorigenesis. Free Radic Biol Med 134:458–467. https://doi.org/10.1016/j.freeradbiomed.2019.01.030
Zhao E, Hou J, Ke X, Abbas MN, Kausar S, Zhang L, Cui H (2019) The roles of sirtuin family proteins in cancer progression. Cancers (Basel) 11(12):1949. https://doi.org/10.3390/cancers11121949
Yang X, Wang Z, Li X, Liu B, Liu M et al (2018) SHMT2 Desuccinylation by SIRT5 Drives Cancer Cell Proliferation. Cancer Res 78(2):372–386. https://doi.org/10.1158/0008-5472.can-17-1912
Wei Z, Song J, Wang G, Cui X, Zheng J et al (2018) Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nat Commun 9(1):4468. https://doi.org/10.1038/s41467-018-06812-y
Zhang Z, Deng X, Liu Y, Liu Y, Sun L (2019) Chen F (2019) PKM2, function and expression and regulation. Cell Biosci 26(9):52. https://doi.org/10.1186/s13578-019-0317-8
Zahra K, Dey T, Ashish MSP, Pandey, (2020) Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Front Oncol 10:159. https://doi.org/10.3389/fonc.2020.00159
Li Z, Yang P (1846) Li Z (2014) The multifaceted regulation and functions of PKM2 in tumor progression. Biochim Biophys Acta 2:285–296. https://doi.org/10.1016/j.bbcan.2014.07.008
Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK et al (2011) Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334(6060):1278–1283. https://doi.org/10.1126/science.1211485
Gui DY, Lewis CA, Vander Heiden MG (2013) Allosteric regulation of PKM2 allows cellular adaptation to different physiological states. Sci Signal 6(263):pe7. https://doi.org/10.1126/scisignal.2003925
Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, Jiang F (2016) PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol Lett 11(3):1980–1986. https://doi.org/10.3892/ol.2016.4168
Marrocco I, Altieri F, Rubini E, Paglia G, Chichiarelli S et al (2019) SHMT2: A STAT3 Signaling new player in prostate cancer energy metabolism. Cells 8(9):1048. https://doi.org/10.3390/cells8091048
Yang P, Li Z, Fu R, Wu H, Li Z (2014) Pyruvate kinase M2 facilitates colon cancer cell migration via the modulation of STAT3 signalling. Cell Signal 26(9):1853–1862. https://doi.org/10.1016/j.cellsig.2014.03.020
Demaria M, Pol V (2012) PKM2, STAT3 and HIF-1α: The Warburg’s vicious circle. JAKSTAT 1(3):194–196. https://doi.org/10.4161/jkst.20662
Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL (2011) Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145(5):732–744. https://doi.org/10.1016/j.cell.2011.03.054
Min DJ, Vural S, Krushkal J (2019) Association of transcriptional levels of folate-mediated one-carbon metabolism-related genes in cancer cell lines with drug treatment response. Cancer Genet 237:19–38. https://doi.org/10.1016/j.cancergen.2019.05.005
Deblois G, Smith HW, Tam IS, Gravel SP, Caron M et al (2016) ERRα mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat Commun 7:12156. https://doi.org/10.1038/ncomms12156
Li X, Zhang K, Hu Y, Luo N (2020) ERRα activates SHMT2 transcription to enhance the resistance of breast cancer to lapatinib via modulating the mitochondrial metabolic adaption. Biosci Rep 40(1):BSR20192465. https://doi.org/10.1042/bsr20192465
Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM (2016) The integrated stress response. EMBO Rep 17(10):1374–1395. https://doi.org/10.15252/embr.201642195
Darini C, Ghaddar N, Chabot C, Assaker G, Sabri S et al (2019) An integrated stress response via PKR suppresses HER2+ cancers and improves trastuzumab therapy. Nat Commun 10(1):2139. https://doi.org/10.1038/s41467-019-10138-8
Wang SF, Wung CH, Chen MS, Chen CF, Yin PH et al (2018) Activated integrated stress response induced by salubrinal promotes cisplatin resistance in human gastric cancer cells via enhanced xCT expression and glutathione biosynthesis. Int J Mol Sci 19(11):3389. https://doi.org/10.3390/ijms19113389
Palam LR, Gore J, Craven KE, Wilson JL, Korc M (2015) Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis 6(10):e1913. https://doi.org/10.1038/cddis.2015.264
Chen L, He J, Zhou J, Xiao Z, Ding N, Duan Y, Li W, Sun LQ (2019) EIF2A promotes cell survival during paclitaxel treatment in vitro and in vivo. J Cell Mol Med 23(9):6060–6071. https://doi.org/10.1111/jcmm.14469
Sethy C, Kundu CN (2021) 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: Implication of DNA repair inhibition. Biomed Pharmacother 137:111285. https://doi.org/10.1016/j.biopha.2021.111285
Rahmani F, Amerizadeh F, Hassanian SM, Hashemzehi M, Nasiri SN et al (2019) PNU-74654 enhances the antiproliferative effects of 5-FU in breast cancer and antagonizes thrombin-induced cell growth via the Wnt pathway. J Cell Physiol 234(8):14123–14132. https://doi.org/10.1002/jcp.28104
Labianca R, Pessi A, Facendola G, Pirovano M, Luporini G (1996) Modulated 5-fluorouracil (5-FU) regimens in advanced colorectal cancer: a critical review of comparative studies. Eur J Cancer 32A(Suppl 5):S7–S12. https://doi.org/10.1016/s0959-8049(96)00330-9
Chen J, Na R, Xiao C, Wang X, Wang Y et al (2021) The loss of SHMT2 mediates 5-fluorouracil chemoresistance in colorectal cancer by upregulating autophagy. Oncogene 40(23):3974–3988. https://doi.org/10.1038/s41388-021-01815-4
Funding
This work was supported by the National Natural Science Foundation of China (No. 81872080, 81572349), Jiangsu Provincial Medical Talent (ZDRCA2016055), the Science and Technology Department of Jiangsu Province (BK20181148), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and the 333 high-level talents of Jiangsu Province (BRA2019083).
Author information
Authors and Affiliations
Contributions
Min Xie wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
For this type of study, informed consent is not required.
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xie, M., Pei, DS. Serine hydroxymethyltransferase 2: a novel target for human cancer therapy. Invest New Drugs 39, 1671–1681 (2021). https://doi.org/10.1007/s10637-021-01144-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-021-01144-z