Abstract
A quantitative RT real-time PCR method was developed for the detection and quantification of Tomato mosaic virus (ToMV) in irrigation waters. These have rarely been monitored for the presence of plant pathogenic viruses, mostly due to the lack of efficient and sensitive detection methods. The newly developed method presented here offers a novel approach in monitoring the health status of environmental waters. ToMV was reliably detected at as low as 12 viral particles per real-time PCR reaction, which corresponds to the initial concentration of approximately 4.2 × 10−10 mg (6,300 viral particles) of ToMV per ml of sample. The sensitivity of the method was further improved by including the Convective Interaction Media® (CIM) monolithic chromatographic columns for quick and efficient concentration of original water samples. Seven out of nine water sources from different locations in Slovenia tested positive for ToMV, after concentrating the sample. Four samples tested ToMV-positive without the concentrating procedure. The presence and integrity of infective ToMV particles in the original sample, as well as in the chromatographic fraction, was confirmed using different methods from test plants, DAS ELISA to electron microscopy and real-time PCR. In this study, we propose a unique and simple diagnostic scheme for rapid, efficient, and sensitive monitoring of irrigation waters that could also be adopted for other plant, human or animal viruses.
Similar content being viewed by others
Introduction
Water quality management is the subject of intense concern. Studies so far have been focused on determining the micro sanitary state of different water resources that could have an impact on human health (Monpoeho et al., 2000; Katayama, Shimasaki, & Ohgaki, 2002; Ho, Chow, & Yau, 2003; Ashbolt, 2004; Pusch et al., 2005). The presence of plant pathogens in water has rarely been monitored (Koenig, 1986; Horvath, Pocsai, & Kazinczi, 1999; Gosalves et al., 2003), mostly due to the lack of efficient and sensitive detection methods. Stricter control is needed for irrigation water and water used in hydroponic cultures (Gosalves et al., 2003), as they may represent a source of infection for plants and consequently crop loss. Viruses present in water can infect plants through the root system and cause the appearance of disease symptoms. They can also be released from infected plants into drainage water and then spread to other plants (Koenig, 1986). Incidences of Melon necrotic spot virus (MNSV) in hydroponic cultures have been reported to have had significant consequences on crops (Gosalves et al., 2003). In a Hungarian study, the presence of 26 plant viruses in 47 environmental water samples was detected (Horvath et al., 1999).
Recently, sensitive real-time PCR methods that enable accurate determination and quantification of pathogens have gained greater acceptance. They have been used to detect and quantify enteroviruses in sludge (Monpoeho et al., 2000), astroviruses in sewage (Le Cann, Ranarijaona, Monpoeho, Le Guyader, & Ferré, 2004), and Cryptosporidium parvum in water samples (Fontaine & Guillot, 2003).
Real-time PCR is also beginning to be used in diagnostics of different plant pathogenic viruses, such as the Potato mop-top virus, Tobacco rattle virus (Boonham, Walsh, Mumford, & Barker, 2000), Tomato spotted wilt virus (Boonham et al., 2002), Barley yellow mosaic virus, Barley mild mosaic virus (Mumford, Skelton, Metcalfe, Walsh, & Boonham, 2004), Cauliflower mosaic virus (Cankar, Ravnikar, Žel, Toplak, & Gruden, 2005), Cucumber vein yellowing virus (Pico, Sifres, & Nuez, 2005), Plum pox virus (Schneider, Sherman, Stone, Damsteegt, & Frederick, 2004; Varga & James, 2005; Olmos, Bertolini, Gil, & Cambra, 2005, Varga & James, 2006), and Potato spindle tuber viroid (Boonham et al., 2004). In each case, real-time PCR has shown significant advantages in detecting disease agents in infected samples, confirming its use as a reliable diagnostic tool. However, no reports have yet been published concerning the use of real-time PCR for the detection of plant pathogenic viruses in environmental waters.
Generally, detection of highly diluted viruses in water samples requires an appropriate concentration step, usually done by PEG precipitation or ultracentrifugation. The latter has been used for astroviruses (Le Cann et al., 2004), whereas a chromatographic concentration method using a negatively charged membrane was developed for the detection of enteroviruses in sea-water (Katayama et al., 2002). Hepatitis A virus has been concentrated from spring water samples using a positively charged membrane (Brassard, Seyer, Houde, Simard, & Trottier, 2005) and CIM QA (Convective Interaction Media®) chromatographic columns were shown to be fast and efficient in the concentration of measles and mumps viruses (Branovic et al., 2003). CIM were also effective in concentrating infectious plant pathogenic ToMV particles, but in dilutions of buffer or tap-water only (Kramberger, Petrovič, Štrancar, & Ravnikar, 2004).
Our aim was to develop a specific real-time PCR for detection and quantification of extremely low concentrations of plant viruses in environmental water samples. For this, we used ToMV, a rod-shaped RNA Tobamovirus of 40 × 106 Da (Caspar, 1963), as our model virus. This method was then adapted for testing environmental water samples and chromatographic fractions, after using CIM chromatographic columns as the concentrating procedure. The above protocol was then employed to assess the health status of several rivers in Slovenia. The relevance of low titres of ToMV as a model for plant health status was also shown using test and propagation plants. The ability to detect infective ToMV in specific natural samples confirms that our newly developed method is rapid, efficient, and highly sensitive. Therefore, it is suitable for evaluating and monitoring irrigation and other water samples.
Materials and methods
Virus purification
ToMV-D strain (provided by Renee Van der Vlugt, PRI, Wageningen, The Netherlands) was used to infect Nicotiana clevelandii propagation plants. The viral particles were isolated from the infected plants by ultracentrifugation in sucrose, followed by caesium chloride gradients, as previously described (Kramberger et al., 2004). Purified ToMV (0.42 mg ml−1) was thus obtained.
Sampling
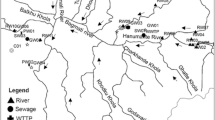
Sampling was performed in late summer 2004, 2005, and 2006 as well as winter 2005. We sampled 2–5 l of water from the following rivers in Slovenia: Krka, Drava, Vipava, and Gradaščica. The same amount was sampled from a water tank in a ToMV-infested greenhouse near Celje, and the same from a gravel pit in Ivanci, located near growing crops. River Krka was sampled in two consecutive years. Soil (1 l) was sampled in the same ToMV-infested greenhouse. It was incubated in 2 l of bi-distilled H2O for 3 days at 4°C. The water was then filtered through filter paper to remove larger particles.
Primer and probe design
Gene sequences of different ToMV isolates were obtained from the public database of the National Centre for Biotechnology Information (NCBI). Multiple sequence alignments of coat protein (CP) genes and movement protein (MP) genes were performed separately using ClustalW 1.8 (BCM search launcher). Since the MP gene sequence was conserved in all the aligned sequences, only one strain was used for designing primers and a probe. Primer Express 2.0 software (Applied Biosystems, Foster City, CA, USA) was used to design primers and a TaqMan® MGB probe to the sequence of the MP gene belonging to ToMV-K strain (accession number AF155507). Primers (ToMVF: 5′-TTG CCG TGG TGG TGT GAG T-3′ starting position at 5154 bp, ToMVR: 5′-GAC CCC AGT GTG GCT TCG T-3′ starting position at 5225 bp) were designed to amplify a 72 bp long amplicon and an MGB probe (ToMVMGB: 5′-TCT TTC CAT TCT CTT GTC AA-3′ starting position at 5202 bp) was designed for specific detection of ToMV. The MGB probe was labelled with FAM (6-carboxyfluorescein) fluorescent dye at the 5′ end and a non-fluorescent quencher and a minor groove binding molecule at the 3′ end.
Reverse transcription and real-time PCR assay
Initial preparation of samples was performed with samples being heated for 10 min at 90°C to break down the possible aggregates of viral particles and to release viral RNA. Samples were diluted 1:10 in 20 mM sodium acetate buffer (pH 5.5) in 1.5 ml tubes. Each sample (20 μl) was pipetted into separate PCR tubes (Applied Biosystems) and denatured by heating at 90°C for 10 min. The reverse transcription mix (20 μl) (cDNA Archive Kit, Applied Biosystems) consisted of: 5.7 μl 10 × PCR buffer (supplied separately, Applied Biosystems), 2.3 μl 25 × dNTP mix, 5.7 μl 1 × random primers, 1.4 μl RNase inhibitor (supplied separately, Applied Biosystems), 2.8 μl Multiscribe reverse transcriptase and 2.1 μl sterile bi-distilled water. It was added to the denatured sample and incubated for 10 min at 25°C and then 120 min at 37°C (GeneAmp® 9700 PCR System, Applied Biosystems).
Real-time PCRs were performed in 20 μl reaction volume with 1 × TaqMan® Universal PCR Master Mix (Applied Biosystems) containing ROX as a passive reference dye for real–time PCR, and AmpErase UNG (uracil n-glycolsylase) to prevent post-PCR contamination.
Concentrations of primers and the probe were optimized in two separate real-time PCR runs. 600 nM primers and 200 nM MGB probe produced the optimal final result which was then used in further experiments. cDNA (4 μl) was then added to the reaction mixture. Real-time PCR reactions were run in triplicate on an ABI PRISM 7900HT Sequence detection system (Applied Biosystem) using universal cycling conditions: 2 min at 50°C, 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. The threshold cycle (Ct) was determined after manual adjustment of the baseline and the fluorescence threshold using SDS 2.2 software (Applied Biosystems). A zero template control (water) and a positive control (purified ToMV in 4.2 × 10−3 mg ml−1) were also included in each run.
The size of RT real-time PCR products was checked on 10% polyacrylamide gel. The PCR product (20 μl) was mixed with 6 μl of loading dye prior to loading onto the gel. To determine the size of the PCR product, 5 μl of ladder (50–2000 bp Ladder, AmpliSize™ Molecular Ruler, Biorad) was mixed with 6 μl of loading dye and also loaded onto the gel. Electrophoresis was run at 4°C and 80 V for 1.5 h; the gel was then stained with ethidium bromide (concentration 10 mg ml−1) for 20 min. UV light was used to visualize the bands.
Determination of the limits of detection, quantification and amplification efficiency
In order to develop a quantitative test, a series of 10-fold dilutions of purified ToMV-D virus particles was prepared. The starting concentration of particles was 0.42 mg ml−1; the series then ranged from 4.2 × 10−3 mg ml−1 to 4.2 × 10−11 mg ml−1 of viral particles. The serial dilutions were assayed by real-time PCR in triplicate in five independent runs. Twenty μl of each viral dilution was then reverse transcribed. Four μl of this cDNA was analyzed by real-time PCR. Standard calibration curves were obtained. This was achieved by plotting Ct values for each reaction against the log of the estimated concentration of viral particles in the sample. The coefficient of linear regression and the slope for the standard curve were then calculated. Such standard curves were used for determining the limits of detection (LOD) and quantification (LOQ). The slope of the standard curve was used to calculate the amplification efficiency according to the equation E = 10(−1/slope)-1, where a value of 1 corresponds to 100% efficiency (Pfaffl, 2001; Ginzinger, 2002).
Detection of ToMV in environmental samples
In order to adapt the above assay for analysis of environmental samples, a series of 10-fold dilutions of ToMV was prepared using irrigation water from the Vipava river, from which ToMV was absent, as shown by the real-time PCR. Concentrations of ToMV in the dilution series ranged from 4.2 × 10−3 mg ml−1 to 4.2 × 10−11 mg ml−1. Samples were assayed in triplicates in four independent real-time PCR runs. The reverse transcription step and the real-time PCR reactions were performed as previously described. Results obtained from dilutions of viral particles in 20 mM sodium acetate buffer and in irrigation water were compared.
We have previously shown that CIM® QA monolithic chromatographic columns can be used to concentrate plant viruses diluted in buffer and tap water (Kramberger et al., 2004). Therefore this same procedure was implemented in the analysis of environmental irrigation water where ToMV could not be directly detected in the sample. Negative samples were concentrated, and elution fractions tested for the presence of ToMV with specific RT real-time PCR as described above.
Concentration of water samples using CIM® QA columns
Three l of water (in which ToMV could not be detected), from the river Gradaščica were divided into three 1 l samples. Following filtration (0.45 μm polyamide filters, Sartorius, Goettingen, Germany) 1 l was spiked with 10 μl and 1 l with 1,000 μl of purified ToMV particles (1.3 × 10−4 mg ml−1), giving final concentrations of 3.31 × 10−9 mg ml−1 and 1.76 × 10−7 mg ml−1, respectively. One l of water was used as a control.
The concentration step was performed as previously described (Kramberger et al., 2004). Eight ml CIM tube monolithic columns were used in order to apply higher flow (40 ml min−1). They were placed in housing and connected to a gradient HPLC system (Knauer, Germany). During the procedure several chromatographic fractions were collected (flow-through, elution, and wash fractions). Twenty mM sodium acetate buffer with sodium chloride (1.5 M) was used to elute the virus from the columns. A ToMV-specific RT real-time PCR reaction was then used to analyse all the samples in triplicate.
Comparison of RT real-time PCR and DAS (double antibody sandwich) ELISA assay
Two methods used for quantification were compared by diluting samples. Extraction buffer was used in DAS ELISA and 20 mM sodium acetate buffer for RT real-time PCR. The sensitivity of each assay was also checked by diluting the ToMV particles in natural irrigation water from the river Vipava. For the DAS ELISA assay, the commercially available antisera (PRI, Wageningen, The Netherlands) were used. Purified viral particles with a known concentration of 0.42 mg ml−1 were diluted in the ELISA extraction buffer as previously reported (Kramberger et al., 2004).
A series of dilutions was made, ranging from 2.5 × 10−5 mg ml−1 to 2.6 × 10−7 mg ml−1 which were then pipetted onto a 96-well ELISA plate (NUNC PolySorp™, Roskilde, Denmark). After 2 h of incubation with substrate (p-nitrophenyl phosphate, Sigma) at a concentration of 1 mg ml−1, absorbance was measured at 405 nm and plotted against the concentration of ToMV in the sample. Logarithmic standard curves were obtained and correlation coefficients calculated. To determine the limit of detection for the DAS ELISA, four independent experiments were compared. Additionally four different environmental water samples (Rivers Drava, Vipava and Krka and water from gravel pit near Ivanci) were tested using both methods. In RT real-time PCR test original and concentrated samples were diluted in 20 mM sodim acetate buffer and in DAS ELISA test in extraction buffer. In RT real-time PCR 20 μl of the original sample was reverse transcribed and 4 μl of obtained cDNA was multiplied in real-time PCR step. 200 μl of the original sample was analysed using the DAS ELISA. After the analyses results were compared and approximate concentrations of ToMV determined.
Infectivity testing
Nicotiana glutinosa test plants were used to test the infectivity of ToMV after the concentration of water samples using CIM. Three leaves per plant were mechanically inoculated with the elution fraction of water sampled from the gravel pit near Ivanci. This sample had been shown to be positive using ToMV-specific RT real-time PCR. Carborundum was put on each leaf and the elution fraction was gently rubbed over it. During the 4 week period following the mechanical inoculation, plants were monitored for the appearance of symptoms and tested for the presence of ToMV using the real-time PCR method. In order to prepare the material for the PCR, plant RNA was isolated using RNeasy Plant Mini Kit (Qiagen, USA). Isolated RNA was then treated with DNase from the DNase I Amplification Grade Kit (Promega, USA) according to the manufacturer’s protocol. ToMV-specific RT real-time PCR was then performed as described above.
Nicotiana clevelandii propagation plants were used to test the infectivity of ToMV in the original sample of irrigation water. Five plants were watered with original water from the gravel pit near Ivanci, where ToMV was present in a concentration of 10−6 mg ml−1. A second set of five plants was watered with a 100-fold dilution of the elution fraction (original concentration of ToMV in the elution fraction was 10−4 mg ml−1), obtained after concentrating the original sample of water from the gravel pit near Ivanci. An additional five plants were used as a negative control and were watered with tap water. After 2 weeks, disease symptoms were monitored and plants were analysed using ToMV-specific real-time PCR.
Electron microscopy
Immuno-serological microscopy (ISEM) was used for the assessment of ToMV particles in the elution fraction following the concentrating procedure of environmental irrigation water, and also for the determination of the presence of ToMV in mechanically inoculated test plants (N. glutinosa). Carbon coated grids were incubated for 45–60 min at room temperature with a 1:1000 dilution of ToMV-specific antibodies (PRI, Wageningen, The Netherlands). The grids were then rinsed with a flow of drops of 0.1 M PBS, pH 7.2 followed by another incubation for 45 min at room temperature with either elution fraction or plant extract. After rinsing, the grids were stained with 6–7 drops of 1% aqueous uranyl acetate and blotted dry. Samples were observed with a Philips CM100 transmission electron microscope.
Results
Design of ToMV-specific quantitative RT real-time PCR
An alignment of sequences of the CP and the MP genes showed that the latter was more conserved than the former sequence, thus making it more suitable for designing a set of primers and a probe which could detect all ToMV viral strains. Assay efficiency and detection limits were compared at the step when RNA was diluted prior to the reverse transcription or when cDNA was diluted after the reverse transcription, as suggested by Stahlberg, Hakansson, Xian, Semb, and Kubista (2004). Diluting RNA prior to RT resulted in greater efficiencies of multiplication in real-time PCR compared to the diluted cDNA (data not shown). Rafikova et al. (2003) observed the formation of ToMV coat protein aggregates at 57°C. In the case of Tobacco mosaic virus (TMV), it was suggested that aggregation can also be controlled by changing temperature. Considering the suggested thermal inactivation point of viral particles (Huttinga and Rast, 1985), the denaturation temperature was set to 90°C for 10 min, which afterwards enabled a more accurate dilution of RNA released from the ToMV particles.
Further experiments showed that the RNA isolation procedure using the RNeasy Plant Mini Kit resulted in a 75% yield (data not shown). This was found by isolating luciferase RNA with a known starting concentration (1 ng μl−1). A comparison was then made between this kit and the above method of RNA release by denaturation at 90°C. Efficiencies and limits of detection of the real-time PCR were assessed, and no significant differences were observed between the two methods. Therefore the less time-consuming temperature denaturation was used in further experiments. In addition to the release of viral RNA, this denaturation step was also performed in order to break the RNA’s secondary structures before the RT step. The formation of such secondary structures has been shown in the case of TMV (Kuznetsov, Daijogo, Zhou, Semler, & McPherson, 2005), also a member of the genus Tobamovirus.
The RT real-time PCR assay was also tested on samples with known concentrations of viral particles, thus providing a calibration curve. Each dilution was tested in triplicate in five independent runs. We considered one typical real-time PCR run (intra-assay) and also compared results between the five independent real-time PCR runs (inter-assay) (Table 1).
In the comparison of the five independent runs, ToMV was detected in all but one dilution tested. The dilution that gave a negative result in all five runs was 4.2 × 10−11 mg of ToMV ml−1 of buffer. When diluting ToMV to a concentration of 4.2 × 10−10 mg ml−1, three of five runs tested positive. The limit of detection (LOD) for real-time PCR was therefore 4.2 × 10−10 mg ml−1. According to Burns Valdivia, and Harris (2004), the concentration value that presents the LOD is where the coefficient of variation (CV) value drops below 50%, and the LOQ is set at a threshold of a CV below 30%. Thus the LOQ for the ToMV-specific real-time PCR was set to 4.2 × 10−8 mg ml−1. Water as a negative control was used in all runs and always tested negative. The slope of the linear regression line was used to calculate the efficiency of the real-time PCR in each individual run. It resulted in efficiencies above 0.84 with mean efficiency of 0.90. Square regression coefficients (R2) of linear regression lines were above 0.987 and the gradients of the slopes ranged from −3.33 to −3.78.
The size of RT real-time PCR bands was checked by analysis of RT real-time PCR products on 10% polyacrylamide gels. The amplification products, visualized on the gel corresponded to the expected size of 72 bp. No difference in the specific band size was observed between the two sample types (one diluted in 20 mM sodium acetate buffer and the other in natural irrigation water). There was also no difference between the specific real-time PCR products run with and without the specific MGB probe. Notably, a faint band at 250 bp was observed only in the sample diluted in 20 mM sodium acetate buffer run without the specific MGB probe. However, as a specific probe was employed rather than a non-specific dsDNA detection system (eg. SYBR®Green), this product did not interfere with the result.
Adaptation of the assay for testing environmental samples
Diluting the virus in irrigation water (from the Vipava river) lowered the sensitivity of the test. Comparison of four independent runs showed that ToMV particles could only be detected at concentrations above 4.2 × 10−5 mg ml−1 and that the concentration 4.2 × 10−4 mg ml−1 presented the limit of quantification. Diluting ToMV in irrigation water also resulted in higher coefficients of variation, ranging from 6.4 to 3.5% when comparing four independent runs. The gradients of the slopes of linear regression lines ranged from −3.54 to −6.28 with square regression coefficients above 0.835. Efficiencies were above 0.44 and mean efficiency was 0.64. For reliable analysis of environmental samples of irrigation water using RT real-time PCR it was necessary to prepare 10-fold dilutions of the original sample in 20 mM sodium acetate buffer. Efficiencies of multiplication improved to the average of 0.90 and the LOD decreased to 4.2 × 10−10 mg ml−1 when buffer was used as a diluent. Also, coefficients of variation were more consistent and the LOQ was lower, reaching 4.2 × 10−8 mg ml−1 (Table 1).
Concentration of environmental samples using CIM monolith chromatographic supports
In order to adapt the previously described concentrating procedure of ToMV particles in tap water using CIM monolithic chromatographic supports, environmental irrigation water from the river Gradaščica was spiked with different amounts of ToMV. Samples were analysed using RT real-time PCR before and after the concentrating procedure with CIM® QA monolithic chromatographic supports. Unspiked water, which was shown to be ToMV-negative even after the concentration step, was employed as a negative control. A sample of water with a ToMV concentration of 3.31 × 10−9 mg ml−1 gave, after CIM chromatography, a concentration greater by two orders of magnitude, i.e. 2.99 × 10−7 mg ml−1. Similarly, a sample with an initial ToMV concentration of 1.76 × 10−7 mg ml−1 also increased in ToMV concentration by two orders of magnitude (to 2.45 × 10−5 mg ml−1) following the concentrating procedure.
Analysis of environmental samples
Different environmental samples were analyzed using RT real-time PCR, before and after the concentrating procedure using CIM monolithic supports. After the concentration procedure, several chromatographic fractions (flow-through, elution, and wash) were obtained and all were tested for the presence of ToMV. Viral particles could be directly detected in four samples of irrigation water; one from a ToMV-infested greenhouse in Celje (tomato plants and soil both also tested ToMV-positive), two samples from the river Drava (sampled in 2004 and 2006), and the fourth from the water sample from the gravel pit in Ivanci sampled in 2005. After the concentrating procedure, three additional samples (elution fraction) tested positive; two samples from the river Krka, taken in two consecutive years and a sample from river Vipava. A sample from the river Gradaščica and a sample from the gravel pit in Ivanci in 2006 both tested negative before and after the concentrating procedure (Table 2).
As can be seen in Table 2, the difference between ToMV starting concentrations and post-CIM concentrations can range from one order of magnitude (in the river Drava) to above six orders of magnitude (in the river Krka – where 10−10 mg ml−1 represents an estimate of the starting concentration). Thus, the efficiency of CIM in concentrating environmental samples may differ depending on the sample. A sample of soil from ToMV-infested greenhouse also tested ToMV-positive with real-time PCR. The sample contained 5 × 10−7 mg ml−1 of ToMV.
Infectivity testing
The infectivity of viral particles was tested after the concentrating procedure. Using ISEM ToMV particles were visualized, and these showed no signs of degradation. Analyses of mechanically inoculated N. glutinosa test plants, where two inoculated leaves and one upper, non-inoculated leaf were tested, showed the presence of ToMV. The concentration of ToMV was estimated to be in the range of 1.6 × 10−8–2.4 × 10−7 mg ml−1 in the inoculated leaves and up to 3.0 × 10−9 mg ml−1 in the upper non-inoculated leaf. In the experiment where three sets of five N. clevelandii propagation plants were watered with ToMV-infested water, the disease symptoms of chlorosis and curling of younger leaves were seen after two weeks. Plant tissue was analysed using quantitative ToMV-specific real-time PCR. The plants which had been watered with a diluted elution fraction tested ToMV-positive with an estimate concentration of 4.0 × 10−9 mg ml−1. The concentration of ToMV in plants watered with original irrigation water from the gravel pit in Ivanci was estimated to be at the limit of detection (LOD) for the method: 4.2 × 10−10 mg ml−1. The plants used as a control had Ct values below those corresponding to our LOD in the test.
Comparison of two ToMV quantification methods
The quantification of ToMV by DAS ELISA (Kramberger et al., 2004) was compared with the newly developed real-time PCR. Diluted samples were analysed and ToMV could be detected in concentrations down to 4.2 × 10−10 mg ml−1 using RT real-time PCR, but only down to 3.1 × 10−5 mg ml−1 using DAS ELISA. The molecular method was thus three orders of magnitude more sensitive than the serological method when comparing the limits of quantification for both methods (LOQ in Table 1), and five orders of magnitude more powerful when comparing limits of detection (LOD in Table 1).
Diluting ToMV in environmental irrigation water lowered the sensitivity of the real-time PCR assay by five orders of magnitude whereas the limit of detection for the DAS ELISA test remained in the same range (10−5 mg ml−1); 4.2 × 10−5 mg ml−1 of ToMV was reliably detected and quantified by real-time PCR and 3.1 × 10−5 mg ml−1 using DAS ELISA. However, when additional dilutions of samples were made in 20 mM sodium acetate buffer, the sensitivity of the molecular method increased to 10−9 mg ml−1. DAS ELISA showed no such deviations, since the sensitivity of the test did not differ.
Four different samples of environmental water were analysed using both methods before and after the concentrating procedure (Table 3). The RT real-time PCR method proved to be more effective, being able to detect the presence of ToMV in two original samples that tested negative using the DAS ELISA method (rivers Drava and Krka). Concentration of ToMV in Drava and Vipava original samples was 10−8 mg ml−1. In concentrated samples ToMV was detected in elution fractions of rivers Vipava and Drava, where concentrations of ToMV, determined using RT real-time PCR ranged from 10−6 mg ml−1 to 10−7 mg ml−1, respectively. Concentrations, determined by semiquantitative DAS ELISA assay were at the LOD and LOQ for the assay – 10−5 mg ml−1.
Discussion
Analysis of environmental irrigation water and the use of CIM monolithic chromatographic supports
The quantitative ToMV-specific RT real-time PCR was developed to monitor the health status of environmental waters, to assess the concentration procedure using CIM, and to compare the sensitivities of RT real-time PCR and DAS ELISA. When adapting the method for testing of environmental irrigation water, there appeared to be a decrease in sensitivity of the test. However, diluting the viral particles in 20 mM sodium acetate buffer improved the sensitivity of the test, the LOD being 4.2 × 10−10 mg ToMV ml−1, which corresponds to 12 ToMV particles per real-time PCR reaction. The size of ToMV particles – 40 × 106 Da (Caspar, 1963) was used to calculate the amount of viral particles where 4.2 × 10−10 mg ToMV per ml corresponds to approximately 6300 ToMV particles ml−1. The volume of all real-time PCR reactions was 20 μl, meaning that as low as 12 viral particles could be detected.
In our experiment, ToMV could not be detected in all environmental samples (Table 2), therefore the concentrating procedure using CIM monolithic chromatographic columns was introduced. CIM had previously been effective in concentrating the virus diluted in buffer and tap water below the detection limit of ELISA (Kramberger et al., 2004). The elution from the CIM was performed using 20 mM sodium acetate buffer with 1.5 M NaCl. However, we diminished the effect of salt as an inhibitor of the PCR by incorporating the denaturation step before diluting the particles in 20 mM sodium acetate buffer. Further analyses of environmental samples showed (Table 2) that CIM is effective in concentrating samples which had shown ToMV concentration levels below the sensitivity of RT real-time PCR. This method was able to concentrate the samples by more than two orders of magnitude – and up to six orders of magnitude. This agrees with our previous findings where CIM treatments were effective in concentrating samples by two to three orders of magnitude (Kramberger et al., 2004). We have shown that ToMV particles were present in the samples from the river Krka (in both 2004 and 2005) and in the river Vipava (in 2006), but could only be detected after the concentration step. The efficiency of CIM to concentrate highly diluted samples has also been demonstrated on measles and mumps viruses (Branovic et al., 2003) where concentration of the samples was increased by 4-fold. Detection of noroviruses in treated sewage has employed RT-PCR, following a two-phase separation concentration method that took two days to complete. RNA was then extracted from the concentrated sample (van den Berg et al., 2005). The concentration method that has been employed in the case of Hepatits A virus and rotaviruses (Brassard et al., 2005) is a combination of adsorption of viruses on positively charged filters, then an elution from the filters, and finally a re-concentration of the eluate. A modified RNA extraction procedure followed, and the analysis of sample was carried out using RT-PCR and Southern blot. It would appear that all the authors proposing new viral detection methods are essentially introducing the same experimental format: concentration then analysis. The main difference seen between the various methods is the time spent from initial sampling to obtaining the final result. In the case of CIM-concentration to detect ToMV in environmental irrigation waters, the full procedure can be run in one day only due to the ability of CIM to sustain higher flows through the chromatographic column and thus contributing to the quicker (less than 30 min) performance of the concentrating procedure.
Furthermore, in our experiments reported here, both ELISA and test plants were also evaluated in order to have three methods available for virus detection that depend on different properties of viral particles. Two different types of test plants were used to assess the infectivity of the ToMV from different water samples: N. glutinosa test plants and N. clevelandii propagation plants. In the latter, ToMV is able to multiply and causes appearance of systemic symptoms, whereas in the former, local symptoms appear as a quick response to the viral infection (Huttinga and Rast, 1985). The infectivity experiments showed that ToMV can be detected in N. glutinosa test plants when they are mechanically inoculated with ToMV-positive elution fraction. This not only suggests the viability of the virus in the original sample, but also its ability to survive the harsh chromatographic conditions, as was additionally shown by electron microscopy. The viability of ToMV was supported by analysis of N. clevelandii propagation plants watered with original water samples. The plants developed typical disease symptoms and ToMV particles could be detected in the plants. The concentration of ToMV in the plants watered with original water was lower than in plants watered with the diluted elution fraction. In the field, however, plants grow for several months in a larger amount of infected water, thus potentially leading to a greater accumulation of plant pathogenic viruses in the host plants.
The molecular detection method was chosen over the previously widely used DAS ELISA following the results of our direct comparison of the two methods. The serological detection method was performed according to the already established diagnostic procedure using polyclonal antibodies (Kramberger et al., 2004). Since the protocol was optimised in the past, the improvement of the serological detection method with monoclonal antibodies was not considered. Duarte, Gomes, Gesztesi, Lopes, and Tavares (2001) who obtained monoclonal antibodies against ToMV reported the low efficiency of such antibody production where only four possible monoclonal antibodies could be produced in low concentrations. Our developed RT real-time PCR was shown to be five orders of magnitude more sensitive than the serological method. This agreed with previous findings (Mumford et al., 2004) where the sensitivities of real-time PCR and ELISA were compared in the cases of Barley yellow mosaic virus and Barley mild mosaic virus. In both these cases, however, the comparisons were qualitative rather than quantitative. Nevertheless, real-time PCR proved to be more sensitive with 77% (60 of 78) of the samples testing positive, compared to the ELISA where only 49% (38 of 78) tested positive. The authors further showed that the real-time PCR was around 100 times (two orders of magnitude) more sensitive than ELISA. Olmos et al. (2005) report a 1,000 times increase in sensitivity of Plum pox virus specific real-time RT-PCR over DASI-ELISA, which is also the difference that was observed in the case of ToMV when limits of quantification are compared.
Monitoring of environmental water
Our study has shown that the monitoring of water samples using the very sensitive and specific molecular method of RT real-time PCR is both very relevant and practical. It can be used to detect live, viable viruses in irrigation water samples. The first step in an analysis procedure for the presence of ToMV should be direct testing of samples of environmental water using RT real-time PCR. In cases where the result of the direct testing is negative, then we suggest that the sample should be concentrated using CIM chromatographic columns. The chromatographic fractions should then be assessed with RT real-time PCR. The results may be confirmed and extended by using additional tests such as test plants and electron microscopy. Such additional test can show important aspects such as virus integrity or infectivity. Furthermore, the DAS ELISA method can still be employed if the concentration of viruses is high enough.
The developed molecular method was designed for evaluation of water samples where the concentration of the virus in the sample cannot be predicted. By employing two-step RT real-time PCR the possible inhibition of real-time PCR reaction due to the high amounts of the target cDNA sequences was eliminated. Separating both steps enables dilution of cDNA before real-time PCR reaction which requires less time for initial optimisation of the amount of RNA put in the RT reaction. Compared to the one-step RT real-time PCR where the detection method for each virus needs to be carefully optimised and the initial amount of the analysed sample needs to be set carefully, the described protocol can be easily applied for detection of other target organisms. The proposed protocol represents a reliable and quick method for screening environmental water samples for the presence of plant pathogenic viruses and has potential applications for animal and human viruses.
References
Ashbolt, N. J. (2004). Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology, 198, 229–238.
van den Berg, H., Lodder, W., van der Poel, W., Vennema, H., & de Roda Husman, A. M. (2005). Genetic diversity of noroviruses in raw and treated sewage water. Research in Microbiology, 156, 532–540.
Boonham, N., González Pérez, L., Mendez, M. S., Lilia Peralta, E., Blockley, A., Walsh, K., Barker, I., & Mumford, R. A. (2004). Development of real-tuime RT-PCR assay for the detection of Potato spindle tuber viroid. Journal of Virological Methods, 116, 139–146.
Boonham, N., Smith, P., Walsh, K., Tame, J., Morris, J., Spence, N., Bennison, J., & Barker, I. (2002). The detection of Tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (TaqMan). Journal of Virological Methods, 101, 37–48.
Boonham, N., Walsh, K., Mumford, R. A., & Barker, I. (2000). Use of multiplex real-time PCR (TaqMan) for the detection of potato viruses. EPPO Bulletin, 30, 427–430.
Branovic, K., Forcic, D., Ivancic, J., Strancar, A., Barut, M., Kosutic-Gulija, T., Zgorelec, R., & Mazuran, R. (2003). Application of short monolithic coloumns for improved detection of viruses. Journal of Virological Methods, 110, 163–171.
Brassard, J., Seyer, K., Houde, A., Simard, C., & Trottier, Y. L. (2005). Concentration and detection of hepatitis A virus and rotavirus in spring water samples by reverse transcription-PCR. Journal of Virological Methods, 123, 163–169.
Burns, M. J., Valdivia, H., & Harris, N. (2004). Analysis and interpretation of data from real-time PCR trace detection methods using quantitation of GM soya as a model system. Analytical and Bioanalytical Chemistry, 378, 1616–1623.
Cankar, K., Ravnikar, M., Žel, J., Toplak, N., & Gruden, K. (2005). Real-time PCR for detection of Cauliflower mosaic virus to complement the 35S screening assay for genetically modified organisms. Journal of AOAC International, 88(3), 841–822.
Caspar, D. L. (1963) Assembly and stability of Tobacco mosaic virus particles. Advances in Protein Chemistry, 18, 37–121.
Duarte, K. M. R., Gomes, L. H., Gesztesi, J-L, Lopes, J. D., & Tavares, F. C. A. (2001). Monclonal antibodies to identify Tomato mosaic tobamovirus (ToMV). Brazilian Journal of Microbiology, 32, 240–242.
Fontaine, M., & Guillot, E. (2003). An immunomagnetic separation-real-time PCR method for quantification of Cryptosporidium parvum in water samples. Journal of Microbiological Method,s 54, 29–36.
Ginzinger, D. G. (2002). Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Experimental Hematology, 30, 503–512.
Gosalves, B., Navarro, J. A., Lorca, A., Botella, F., Sánchez-Pina, M. A., & Pallas, V. (2003). Detection of Melon necrotic spot virus in water samples and melon plants by molecular methods. Journal of Virological Methods, 113, 87–93.
Ho, K. C., Chow, Y. L., & Yau, J. T. S. (2003). Chemical and microbiological qualities of The East River (Dongjiang) water, with particular reference to drinking water supply in Hong Kong. Chemosphere, 52, 1441–1450.
Horvath, J., Pocsai E., & Kazinczi, G. (1999) Plant virus contamination of natural waters in Hungary. In J. Maček (ed.), Lecture and papers presented at the fourth slovenian conference on plant protection in Portorož, Ljubljana (3–4 March, 1999, pp. 353–356). Ljubljana: Plant Protection Society of Slovenia.
Huttinga, H., & Rast, A. Th. B. (1985) Tomato mosaic tobamovirus. Plant Viruses Online, Descriptions and Lists from the VIDE Database. Available online: http://image.fs.idaho.edu/vide/descr832.htm (9. 3. 2004).
Katayama, H., Shimasaki, A., & Ohgaki, S. (2002). Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Applied and Environmental Microbiology, 68(3), 1033–1039.
Koenig, R. (1986). Plant viruses in rivers and lakes. Advances in Virus Research, 31, 321–333.
Kramberger, P., Petrovič, N., Štrancar, A., & Ravnikar, M. (2004). Concentration of plant viruses using monolithic chromatographic supports. Journal of Virological Methods, 120, 51–57.
Kuznetsov, Y. G., Daijogo, S., Zhou, J., Semler, B. L., & McPherson, A. (2005). Atomic force microscopy analysis of icosahedral virus RNA. Journal of Molecular Biology, 347, 41–52.
Le Cann, P., Ranarijaona, S., Monpoeho, S., Le Guyader, F., & Ferré, V. (2004). Quantification of human astroviruses in sewage using real-time RT-PCR. Research in Microbiology, 155, 11–15.
Monpoeho, S., Dehée, A., Mignotte, B., Schwartzbrod, L., Marechal, V., Nicolas, J. C., Billaudel, S., & Férré, V. (2000). Quantification of Enterovirus RNA in Sludge Samples Using Single Tube Real-Time RT-PCR. BioTechniques, 29, 88–93.
Mumford, R., Skelton, A., Metcalfe, E., Walsh, K., & Boonham, N. (2004). The reliable detection of Barley yellow and mild mosaic viruses using real-time PCR (TaqMan®). Journal of Virological Methods, 117, 153–159.
Olmos, A., Bertolini, E., Gil, M., & Cambra, M. (2005). Real-time assay for quantitative detection of non-persistently transmitted Plum pox virus RNA targets in single aphids. Journal of Virological Methods, 128, 151–155.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time PCR. Nucleic Acids Research, 29(9), 2002–2007.
Pico, B., Sifres, A., & Nuez, F. (2005). Quantitative detection of Cucumber vein yellowing virus in susceptible and partially resistant plants using real-time PCR. Journal of Virological Methods, 128, 14–20.
Pusch, D., Oh, D. Y., Wolf, S., Dumke, R., Schröter-Bobsin, U., Höhne, M., Röske, I., & Schreier, E. (2005). Detection of enteric viruses and bacterial indicators in German environmental waters. Archives of Virology, 150(5), 929–947.
Rafikova, E. R., Kurganov, B. I., Arutyunyan, A. M., Kust, S. V., Drachev, V. A., & Dobrov, E. N. (2003). A mechanism of macroscopic (amorphous) aggregation of the tobacco mosaic virus coat protein. The International Journal of Biochemistry and Cell Biology, 35, 1452–1460.
Schneider, W. L., Sherman, D. J., Stone, A. L., Damsteegt, V. D., & Frederick, R. D. (2004) Specific detection and quantification of Plum pox virus by real-time fluorescent reverse transcription-PCR. Journal of Virological Methods, 120, 97–105.
Stahlberg, A., Hakansson, J., Xian, X., Semb, H., & Kubista, M. (2004). Properties of the reverse transcription reaction in mRNA quantification. Clinical Chemistry, 50(3), 509–515.
Varga, A., & James, D (2005). Detection and differentiation of Plum pox virus using real-time multiplex PCR with SYBR Green and melting curve analysis: a rapid method for strain typing. Journal of Virological Methods, 123, 213–220.
Varga, A., & James, D. (2006). Real-time RT-PCR and SYBR Green I melting curve analysis for the identification of Plum pox virus strains C, EA, and W: Effect of amplicon size, melt rate, and dye translocation. Journal of Virological Methods, 132, 146–153.
Acknowledgements
The authors would like to thank the Ministry of Higher Education, Science and Technology of Republic of Slovenia for co-financing the project (Ref. No. L4-6050). We are also grateful to Dr. Renee Van der Vlugt from Plant Research International, Wageningen, The Netherlands for providing the ToMV-D strain and the protocol for purifying virus particles, to the Phytosanitary Inspection of Republic of Slovenia and Aleš Blatnik (NIB) for providing us with the water samples, Magda Tušek Žnidarič for examining samples under the electron microscope, Dr. Neil Boonham (CSL, York, UK) for valuable discussions, and Dr. Mike Galsworthy for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Boben, J., Kramberger, P., Petrovič, N. et al. Detection and quantification of Tomato mosaic virus in irrigation waters. Eur J Plant Pathol 118, 59–71 (2007). https://doi.org/10.1007/s10658-007-9112-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-007-9112-1